Mitochondrial biogenesis and turnover
- PMID: 18395251
- PMCID: PMC3175594
- DOI: 10.1016/j.ceca.2007.12.004
Mitochondrial biogenesis and turnover
Abstract
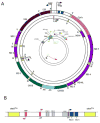
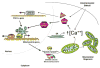
Mitochondrial biogenesis is a complex process involving the coordinated expression of mitochondrial and nuclear genes, the import of the products of the latter into the organelle and turnover. The mechanisms associated with these events have been intensively studied in the last 20 years and our understanding of their details is much improved. Mitochondrial biogenesis requires the participation of calcium signaling that activates a series of calcium-dependent protein kinases that in turn activate transcription factors and coactivators such as PGC-1alpha that regulates the expression of genes coding for mitochondrial components. In addition, mitochondrial biogenesis involves the balance of mitochondrial fission-fusion. Mitochondrial malfunction or defects in any of the many pathways involved in mitochondrial biogenesis can lead to degenerative diseases and possibly play an important part in aging.
Figures

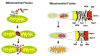
References
-
- DiMauro S, Schon EA. Mitochondrial respiratory-chain diseases. N Engl J Med. 2003;348:2656–68. - PubMed
-
- Wallace D. Mitochondrial defects in neurodegenerative disease. Mental Retardation and Developmental Disabilities Research Reviews. 2001;7:158–166. - PubMed
-
- Margulis L. Symbiotic theory of the origin of eukaryotic organelles; criteria for proof. Symp Soc Exp Biol. 1975:21–38. - PubMed
-
- Margulis L, Bermudes D. Symbiosis as a mechanism of evolution: status of cell symbiosis theory. Symbiosis. 1985;1:101–24. - PubMed
-
- Shutt TE, Gray MW. Bacteriophage origins of mitochondrial replication and transcription proteins. Trends Genet. 2006;22:90–5. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

