Natural selection of protein structural and functional properties: a single nucleotide polymorphism perspective
- PMID: 18397526
- PMCID: PMC2643940
- DOI: 10.1186/gb-2008-9-4-r69
Natural selection of protein structural and functional properties: a single nucleotide polymorphism perspective
Abstract
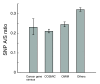
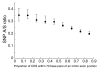
Background: The rates of molecular evolution for protein-coding genes depend on the stringency of functional or structural constraints. The Ka/Ks ratio has been commonly used as an indicator of selective constraints and is typically calculated from interspecies alignments. Recent accumulation of single nucleotide polymorphism (SNP) data has enabled the derivation of Ka/Ks ratios for polymorphism (SNP A/S ratios).
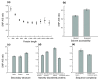
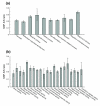
Results: Using data from the dbSNP database, we conducted the first large-scale survey of SNP A/S ratios for different structural and functional properties. We confirmed that the SNP A/S ratio is largely correlated with Ka/Ks for divergence. We observed stronger selective constraints for proteins that have high mRNA expression levels or broad expression patterns, have no paralogs, arose earlier in evolution, have natively disordered regions, are located in cytoplasm and nucleus, or are related to human diseases. On the residue level, we found higher degrees of variation for residues that are exposed to solvent, are in a loop conformation, natively disordered regions or low complexity regions, or are in the signal peptides of secreted proteins. Our analysis also revealed that histones and protein kinases are among the protein families that are under the strongest selective constraints, whereas olfactory and taste receptors are among the most variable groups.
Conclusion: Our study suggests that the SNP A/S ratio is a robust measure for selective constraints. The correlations between SNP A/S ratios and other variables provide valuable insights into the natural selection of various structural or functional properties, particularly for human-specific genes and constraints within the human lineage.
Figures




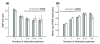
References
-
- Li W-H. Molecular Evolution. Sunderland, Massachusetts: Sinauer Associates, Inc.; 1997.
-
- Mouse Genome Sequencing Consortium. Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, Antonarakis SE, Attwood J, Baertsch R, Bailey J, Barlow K, Beck S, Berry E, Birren B, Bloom T, Bork P, Botcherby M, Bray N, Brent MR, Brown DG, Brown SD, Bult C, Burton J, Butler J, Campbell RD, Carninci P. et al.Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

