Molecular signature of hypersaline adaptation: insights from genome and proteome composition of halophilic prokaryotes
- PMID: 18397532
- PMCID: PMC2643941
- DOI: 10.1186/gb-2008-9-4-r70
Molecular signature of hypersaline adaptation: insights from genome and proteome composition of halophilic prokaryotes
Abstract
Background: Halophilic prokaryotes are adapted to thrive in extreme conditions of salinity. Identification and analysis of distinct macromolecular characteristics of halophiles provide insight into the factors responsible for their adaptation to high-salt environments. The current report presents an extensive and systematic comparative analysis of genome and proteome composition of halophilic and non-halophilic microorganisms, with a view to identify such macromolecular signatures of haloadaptation.
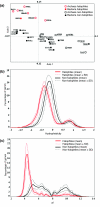
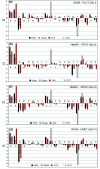
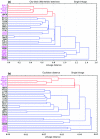
Results: Comparative analysis of the genomes and proteomes of halophiles and non-halophiles reveals some common trends in halophiles that transcend the boundary of phylogenetic relationship and the genomic GC-content of the species. At the protein level, halophilic species are characterized by low hydrophobicity, over-representation of acidic residues, especially Asp, under-representation of Cys, lower propensities for helix formation and higher propensities for coil structure. At the DNA level, the dinucleotide abundance profiles of halophilic genomes bear some common characteristics, which are quite distinct from those of non-halophiles, and hence may be regarded as specific genomic signatures for salt-adaptation. The synonymous codon usage in halophiles also exhibits similar patterns regardless of their long-term evolutionary history.
Conclusion: The generality of molecular signatures for environmental adaptation of extreme salt-loving organisms, demonstrated in the present study, advocates the convergent evolution of halophilic species towards specific genome and amino acid composition, irrespective of their varying GC-bias and widely disparate taxonomic positions. The adapted features of halophiles seem to be related to physical principles governing DNA and protein stability, in response to the extreme environmental conditions under which they thrive.
Figures




References
-
- Eisenberg H, Mevarech M, Zaccai G. Biochemical, structural, and molecular genetic aspects of halophilism. Adv Protein Chem. 1992;43:1–62. - PubMed
-
- Galinsky EA, Trüper HG. Microbial behaviour in salt-stressed ecosystems. FEMS Microbiol Rev. 1994;15:95–108. doi: 10.1111/j.1574-6976.1994.tb00128.x. - DOI
-
- Mongodin EF, Nelson KE, Daugherty S, Deboy RT, Wister J, Khouri H, Weidman J, Walsh DA, Papke RT, Sanchez Perez G, Sharma AK, Nesbo CL, MacLeod D, Bapteste E, Doolittle WF, Charlebois RL, Legault B, Rodriguez-Valera F. The genome of Salinibacter ruber : convergence and gene exchange among hyperhalophilic bacteria and archaea. Proc Natl Acad Sci USA. 2005;102:18147–18152. doi: 10.1073/pnas.0509073102. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

