Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi
- PMID: 18398004
- PMCID: PMC2311380
- DOI: 10.1073/pnas.0801775105
Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi
Abstract
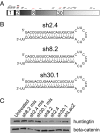
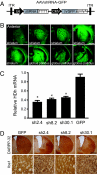
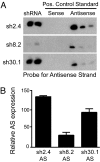
Huntington's disease (HD) is a fatal, dominant neurodegenerative disease caused by a polyglutamine repeat expansion in exon 1 of the HD gene, which encodes the huntingtin protein. We and others have shown that RNAi is a candidate therapy for HD because expression of inhibitory RNAs targeting mutant human HD transgenes improved neuropathology and behavioral deficits in HD mouse models. Here, we developed shRNAs targeting conserved sequences in human HD and mouse HD homolog (HDh) mRNAs to initiate preclinical testing in a knockin mouse model of HD. We screened 35 shRNAs in vitro and subsequently narrowed our focus to three candidates for in vivo testing. Unexpectedly, two active shRNAs induced significant neurotoxicity in mouse striatum, although HDh mRNA expression was reduced to similar levels by all three. Additionally, a control shRNA containing mismatches also induced toxicity, although it did not reduce HDh mRNA expression. Interestingly, the toxic shRNAs generated higher antisense RNA levels, compared with the nontoxic shRNA. These results demonstrate that the robust levels of antisense RNAs emerging from shRNA expression systems can be problematic in the mouse brain. Importantly, when sequences that were toxic in the context of shRNAs were placed into artificial microRNA (miRNA) expression systems, molecular and neuropathological readouts of neurotoxicity were significantly attenuated without compromising mouse HDh silencing efficacy. Thus, miRNA-based approaches may provide more appropriate biological tools for expressing inhibitory RNAs in the brain, the implications of which are crucial to the development of RNAi for both basic biological and therapeutic applications.
Conflict of interest statement
Conflict of interest statement: B.L.D. was a consultant for Sirna Therapeutics, Inc.
Figures
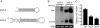
References
-
- Fire A, et al. Potent and specific genetic interference by double-stranded RNA in caenorhabditis elegans. Nature. 1998;391:806–811. - PubMed
-
- Reinhart BJ, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. - PubMed
-
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. - PubMed
-
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. - PubMed
-
- Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

