GPR50 is the mammalian ortholog of Mel1c: evidence of rapid evolution in mammals
- PMID: 18400093
- PMCID: PMC2323367
- DOI: 10.1186/1471-2148-8-105
GPR50 is the mammalian ortholog of Mel1c: evidence of rapid evolution in mammals
Erratum in
- BMC Evol Biol. 2012;12:28
Abstract
Background: The melatonin receptor subfamily contains three members Mel1a, Mel1b and Mel1c, found in all vertebrates except for Mel1c which is found only in fish, Xenopus species and the chicken. Another receptor, the melatonin related receptor known as GPR50, found exclusively in mammals and later identified as a member of the melatonin receptor subfamily because of its identity to the three melatonin receptors despite its absence of affinity for melatonin. The aim of this study was to describe the evolutionary relationships between GPR50 and the three other members of the melatonin receptor subfamily.
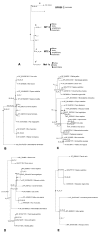
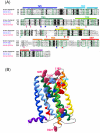
Results: Using an in silico approach, we demonstrated that GPR50 is the ortholog of the high affinity Mel1c receptor. It was necessary to also study the synteny of this gene to reach this conclusion because classical mathematical models that estimate orthology and build phylogenetic trees were not sufficient. The receptor has been deeply remodelled through evolution by the mutation of numerous amino acids and by the addition of a long C-terminal tail. These alterations have modified its affinity for melatonin and probably affected its interactions with the other two known melatonin receptors MT1 and MT2 that are encoded by Mel1a and Mel1b genes respectively. Evolutionary studies provided evidence that the GPR50 group evolved under different selective pressure as compared to the orthologous groups Me11 a, b, and c.
Conclusion: This study demonstrated that there are only three members in the melatonin receptor subfamily with one of them (Me11c) undergoing rapid evolution from fishes and birds to mammals. Further studies are necessary to investigate the physiological roles of this receptor.
Figures




References
-
- Audinot V, Mailliet F, Lahaye-Brasseur C, Bonnaud A, Le Gall A, Amosse C, Dromaint S, Rodriguez M, Nagel N, Galizzi JP, Malpaux B, Guillaumet G, Lesieur D, Lefoulon F, Renard P, Delagrange P, Boutin JA. New selective ligands of human cloned melatonin MT1 and MT2 receptors. Naunyn Schmiedebergs Arch Pharmacol. 2003;367(6):553–561. doi: 10.1007/s00210-003-0751-2. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

