Site 1 protease is required for proteolytic processing of the glycoproteins of the South American hemorrhagic fever viruses Junin, Machupo, and Guanarito
- PMID: 18400865
- PMCID: PMC2395157
- DOI: 10.1128/JVI.02392-07
Site 1 protease is required for proteolytic processing of the glycoproteins of the South American hemorrhagic fever viruses Junin, Machupo, and Guanarito
Abstract
The cellular proprotein convertase site 1 protease (S1P) has been implicated in the proteolytic processing of the glycoproteins (GPs) of Old World arenaviruses. Here we report that S1P is also involved in the processing of the GPs of the genetically more-distant South American hemorrhagic fever viruses Guanarito, Machupo, and Junin. Efficient cleavage of Guanarito virus GP, whose protease recognition sites deviate from the reported S1P consensus sequence, indicates a broader specificity of S1P than anticipated. Lack of GP processing of Junin virus dramatically reduced production of infectious virus and prevented cell-to-cell propagation. Infection of S1P-deficient cells resulted in viral persistence over several weeks without the emergence of escape variants able to use other cellular proteases for GP processing.
Figures


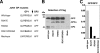
References
-
- Brown, M. S., and J. L. Goldstein. 1997. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89331-340. - PubMed
-
- Buchmeier, M. J., J. C. de la Torre, and C. J. Peters. 2007. Arenaviridae: the viruses and their replication, p. 1791-1828. In D. L. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott-Raven, Philadelphia, PA.
-
- Enria, D. A., and J. G. Barrera Oro. 2002. Junin virus vaccines. Curr. Top. Microbiol. Immunol. 263239-261. - PubMed
-
- Geisbert, T. W., and P. B. Jahrling. 2004. Exotic emerging viral diseases: progress and challenges. Nat. Med. 10S110-S1121. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

