Cleavage of group 1 coronavirus spike proteins: how furin cleavage is traded off against heparan sulfate binding upon cell culture adaptation
- PMID: 18400867
- PMCID: PMC2395124
- DOI: 10.1128/JVI.00074-08
Cleavage of group 1 coronavirus spike proteins: how furin cleavage is traded off against heparan sulfate binding upon cell culture adaptation
Abstract
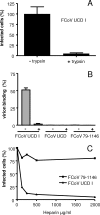
A longstanding enigmatic feature of the group 1 coronaviruses is the uncleaved phenotype of their spike protein, an exceptional property among class I fusion proteins. Here, however, we show that some group 1 coronavirus spike proteins carry a furin enzyme recognition motif and can actually be cleaved, as demonstrated for a feline coronavirus. Interestingly, this feature can be lost during cell culture adaptation by a single mutation in the cleavage motif; this, however, preserves a heparan sulfate binding motif and renders infection by the virus heparan sulfate dependent. We identified a similar cell culture adaptation for the human coronavirus OC43.
Figures


References
-
- Binns, M. M., M. E. Boursnell, D. Cavanagh, D. J. Pappin, and T. D. Brown. 1985. Cloning and sequencing of the gene encoding the spike protein of the coronavirus IBV. J. Gen. Virol. 66719-726. - PubMed
-
- Bosch, B. J., and P. J. Rottier. 2008. Nidovirus entry into cells, p. 157-177. In S. Perlman, T. Gallagher, and E. J. Snijder (ed.), Nidoviruses. ASM Press, Washington, DC.
-
- Cavanagh, D., P. J. Davis, D. J. Pappin, M. M. Binns, M. E. Boursnell, and T. D. Brown. 1986. Coronavirus IBV: partial amino terminal sequencing of spike polypeptide S2 identifies the sequence Arg-Arg-Phe-Arg-Arg at the cleavage site of the spike precursor propolypeptide of IBV strains Beaudette and M41. Virus Res. 4133-143. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

