Quantitative proliferation dynamics and random chromosome segregation of hair follicle stem cells
- PMID: 18401343
- PMCID: PMC2374848
- DOI: 10.1038/emboj.2008.72
Quantitative proliferation dynamics and random chromosome segregation of hair follicle stem cells
Abstract
Regulation of stem cell (SC) proliferation is central to tissue homoeostasis, injury repair, and cancer development. Accumulation of replication errors in SCs is limited by either infrequent division and/or by chromosome sorting to retain preferentially the oldest 'immortal' DNA strand. The frequency of SC divisions and the chromosome-sorting phenomenon are difficult to examine accurately with existing methods. To address this question, we developed a strategy to count divisions of hair follicle (HF) SCs over time, and provide the first quantitative proliferation history of a tissue SC during its normal homoeostasis. We uncovered an unexpectedly high cellular turnover in the SC compartment in one round of activation. Our study provides quantitative data in support of the long-standing infrequent SC division model, and shows that HF SCs do not retain the older DNA strands or sort their chromosome. This new ability to count divisions in vivo has relevance for obtaining basic knowledge of tissue kinetics.
Figures
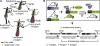
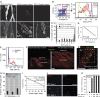
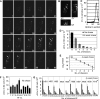



References
-
- Armakolas A, Klar AJ (2006) Cell type regulates selective segregation of mouse chromosome 7 DNA strands in mitosis. Science 311: 1146–1149 - PubMed
-
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H (2007) Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449: 1003–1007 - PubMed
-
- Bickenbach JR, Chism E (1998) Selection and extended growth of murine epidermal SCs in culture. Exp Cell Res 244: 184–195 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

