The three SoxC proteins--Sox4, Sox11 and Sox12--exhibit overlapping expression patterns and molecular properties
- PMID: 18403418
- PMCID: PMC2396431
- DOI: 10.1093/nar/gkn162
The three SoxC proteins--Sox4, Sox11 and Sox12--exhibit overlapping expression patterns and molecular properties
Abstract
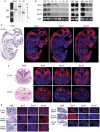
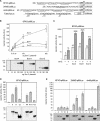
The group C of Sry-related high-mobility group (HMG) box (Sox) transcription factors has three members in most vertebrates: Sox4, Sox11 and Sox12. Sox4 and Sox11 have key roles in cardiac, neuronal and other major developmental processes, but their molecular roles in many lineages and the roles of Sox12 remain largely unknown. We show here that the three genes are co-expressed at high levels in neuronal and mesenchymal tissues in the developing mouse, and at variable relative levels in many other tissues. The three proteins have conserved remarkable identity through evolution in the HMG box DNA-binding domain and in the C-terminal 33 residues, and we demonstrate that the latter residues constitute their transactivation domain (TAD). Sox11 activates transcription several times more efficiently than Sox4 and up to one order of magnitude more efficiently than Sox12, owing to a more stable alpha-helical structure of its TAD. This domain and acidic domains interfere with DNA binding, Sox11 being most affected and Sox4 least affected. The proteins are nevertheless capable of competing with one another in reporter gene transactivation. We conclude that the three SoxC proteins have conserved overlapping expression patterns and molecular properties, and might therefore act in concert to fulfill essential roles in vivo.
Figures







References
-
- Schepers GE, Teasdale RD, Koopman P. Twenty pairs of Sox: extent, homology, and nomenclature of the mouse and human Sox transcription factor gene families. Dev. Cell. 2002;3:167–170. - PubMed
-
- Koopman P, Gubbay J, Vivian N, Goodfellow P, Lovell-Badge R. Male development of chromosomally female mice transgenic for Sry. Nature. 1991;351:117–121. - PubMed
-
- Bianchi ME, Agresti A. HMG proteins: dynamic players in gene regulation and differentiation. Curr. Opin. Genet. Dev. 2005;15:496–506. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

