Evaluation and validation of total RNA extraction methods for microRNA expression analyses in formalin-fixed, paraffin-embedded tissues
- PMID: 18403610
- PMCID: PMC2329784
- DOI: 10.2353/jmoldx.2008.070153
Evaluation and validation of total RNA extraction methods for microRNA expression analyses in formalin-fixed, paraffin-embedded tissues
Abstract
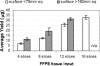
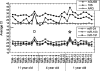
Histopathology archives of well-annotated formalin-fixed, paraffin-embedded (FFPE) tissue specimens are valuable resources for retrospective studies of human diseases. Since recovery of quality intact mRNA compatible with molecular techniques is often difficult due to degradation, we evaluated microRNA (miRNA), a novel class of small RNA molecules with growing therapeutic and diagnostic potential, as an alternative analyte for gene expression studies of FFPE samples. Analyzing total RNA yield, miRNA recovery, and robustness of real-time polymerase chain reaction for miRNA, mRNA, and rRNA species, we compared the performance of commercially available RNA isolation kits and identified a preferred methodology. We further implemented modifications to increase tissue throughput and incorporate a quantitative Armored RNA process control to monitor RNA recovery efficiency. The optimized process was tested for reproducibility as well as interoperator and interday variability, and was validated with a set of 30 clinical samples. In addition, we demonstrated that, independent of FFPE block age and RNA quality, miRNAs generate quantitative reverse transcription-polymerase chain reaction signals that are more robust and better correlate with expression levels from frozen reference samples compared with longer mRNAs. Our broad study, including a total of 272 independent RNA isolations from 17 tissue types and 65 FFPE blocks up to 12 years old, indicates that miRNAs are not only suitable but are also likely superior analytes for the molecular characterization of compromised archived clinical specimens.
Figures


References
-
- Lewis F, Maughan NJ, Smith V, Hillan K, Quirke P. Unlocking the archive–gene expression in paraffin-embedded tissue. J Pathol. 2001;195:66–71. - PubMed
-
- Chung JY, Braunschweig T, Hewitt SM. Optimization of recovery of RNA from formalin-fixed, paraffin-embedded tissue. Diagn Mol Pathol. 2006;15:229–236. - PubMed
-
- Korga A, Wilkolaska K, Korobowicz E. Difficulties in using archival paraffin-embedded tissues for RNA expression analysis. Postepy Hig Med Dosw (online) 2007;61:151–155. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

