A generalized allosteric mechanism for cis-regulated cyclic nucleotide binding domains
- PMID: 18404204
- PMCID: PMC2275311
- DOI: 10.1371/journal.pcbi.1000056
A generalized allosteric mechanism for cis-regulated cyclic nucleotide binding domains
Erratum in
- PLoS Comput Biol. 2009 Jul;5(7). doi: 10.1371/annotation/43fd4ca3-0996-462c-ad89-395c369cbaa2
Abstract
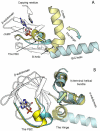
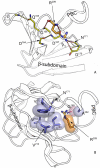
Cyclic nucleotides (cAMP and cGMP) regulate multiple intracellular processes and are thus of a great general interest for molecular and structural biologists. To study the allosteric mechanism of different cyclic nucleotide binding (CNB) domains, we compared cAMP-bound and cAMP-free structures (PKA, Epac, and two ionic channels) using a new bioinformatics method: local spatial pattern alignment. Our analysis highlights four major conserved structural motifs: 1) the phosphate binding cassette (PBC), which binds the cAMP ribose-phosphate, 2) the "hinge," a flexible helix, which contacts the PBC, 3) the beta(2,3) loop, which provides precise positioning of an invariant arginine from the PBC, and 4) a conserved structural element consisting of an N-terminal helix, an eight residue loop and the A-helix (N3A-motif). The PBC and the hinge were included in the previously reported allosteric model, whereas the definition of the beta(2,3) loop and the N3A-motif as conserved elements is novel. The N3A-motif is found in all cis-regulated CNB domains, and we present a model for an allosteric mechanism in these domains. Catabolite gene activator protein (CAP) represents a trans-regulated CNB domain family: it does not contain the N3A-motif, and its long range allosteric interactions are substantially different from the cis-regulated CNB domains.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
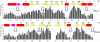

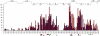


Similar articles
-
Understanding cAMP-dependent allostery by NMR spectroscopy: comparative analysis of the EPAC1 cAMP-binding domain in its apo and cAMP-bound states.J Am Chem Soc. 2007 Nov 21;129(46):14482-92. doi: 10.1021/ja0753703. Epub 2007 Oct 31. J Am Chem Soc. 2007. PMID: 17973384
-
Mutations of PKA cyclic nucleotide-binding domains reveal novel aspects of cyclic nucleotide selectivity.Biochem J. 2017 Jul 6;474(14):2389-2403. doi: 10.1042/BCJ20160969. Biochem J. 2017. PMID: 28583991 Free PMC article.
-
Activation of PKA via asymmetric allosteric coupling of structurally conserved cyclic nucleotide binding domains.Nat Commun. 2019 Sep 4;10(1):3984. doi: 10.1038/s41467-019-11930-2. Nat Commun. 2019. PMID: 31484930 Free PMC article.
-
The structure of the apo cAMP-binding domain of HCN4 - a stepping stone toward understanding the cAMP-dependent modulation of the hyperpolarization-activated cyclic-nucleotide-gated ion channels.FEBS J. 2018 Jun;285(12):2182-2192. doi: 10.1111/febs.14408. Epub 2018 Mar 14. FEBS J. 2018. PMID: 29444387 Review.
-
cAMP-dependent allostery and dynamics in Epac: an NMR view.Biochem Soc Trans. 2012 Feb;40(1):219-23. doi: 10.1042/BST20110628. Biochem Soc Trans. 2012. PMID: 22260694 Review.
Cited by
-
Crystal Structure of PKG I:cGMP Complex Reveals a cGMP-Mediated Dimeric Interface that Facilitates cGMP-Induced Activation.Structure. 2016 May 3;24(5):710-720. doi: 10.1016/j.str.2016.03.009. Epub 2016 Apr 7. Structure. 2016. PMID: 27066748 Free PMC article.
-
Discovery of Allostery in PKA Signaling.Biophys Rev. 2015 Jun 1;7(2):227-238. doi: 10.1007/s12551-015-0170-x. Biophys Rev. 2015. PMID: 26097522 Free PMC article.
-
Structural and evolutionary divergence of cyclic nucleotide binding domains in eukaryotic pathogens: Implications for drug design.Biochim Biophys Acta. 2015 Oct;1854(10 Pt B):1575-85. doi: 10.1016/j.bbapap.2015.03.012. Epub 2015 Apr 3. Biochim Biophys Acta. 2015. PMID: 25847873 Free PMC article.
-
Parallel Allostery by cAMP and PDE Coordinates Activation and Termination Phases in cAMP Signaling.Biophys J. 2015 Sep 15;109(6):1251-63. doi: 10.1016/j.bpj.2015.06.067. Epub 2015 Aug 11. Biophys J. 2015. PMID: 26276689 Free PMC article.
-
Electrostatic Interactions as Mediators in the Allosteric Activation of Protein Kinase A RIα.Biochemistry. 2017 Mar 14;56(10):1536-1545. doi: 10.1021/acs.biochem.6b01152. Epub 2017 Mar 6. Biochemistry. 2017. PMID: 28221775 Free PMC article.
References
-
- Beavo JA, Brunton LL. Cyclic nucleotide research - still expanding after half a century. Nat Rev Mol Cell Biol. 2002;3:710–718. - PubMed
-
- Bos JL. Epac: a new cAMP target and new avenues in cAMP research. Nat Rev Mol Cell Biol. 2003;4:733–738. - PubMed
-
- Kaupp UB, Seifert R. Cyclic nucleotide-gated ion channels. Physiol Rev. 2002;82:769–824. - PubMed
-
- Kopperud R, Krakstad C, Selheim F, Doskeland SO. cAMP effector mechanisms. Novel twists for an ‘old’ signaling system. FEBS Lett. 2003;546:121–126. - PubMed
-
- Newton RP, Smith CJ. Cyclic nucleotides. Phytochemistry. 2004;65:2423–2437. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

