Binding-induced folding of a natively unstructured transcription factor
- PMID: 18404207
- PMCID: PMC2289845
- DOI: 10.1371/journal.pcbi.1000060
Binding-induced folding of a natively unstructured transcription factor
Abstract
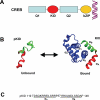
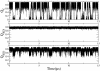
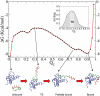
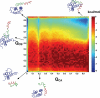
Transcription factors are central components of the intracellular regulatory networks that control gene expression. An increasingly recognized phenomenon among human transcription factors is the formation of structure upon target binding. Here, we study the folding and binding of the pKID domain of CREB to the KIX domain of the co-activator CBP. Our simulations of a topology-based Gō-type model predict a coupled folding and binding mechanism, and the existence of partially bound intermediates. From transition-path and Phi-value analyses, we find that the binding transition state resembles the unstructured state in solution, implying that CREB becomes structured only after committing to binding. A change of structure following binding is reminiscent of an induced-fit mechanism and contrasts with models in which binding occurs to pre-structured conformations that exist in the unbound state at equilibrium. Interestingly, increasing the amount of structure in the unbound pKID reduces the rate of binding, suggesting a "fly-casting"-like process. We find that the inclusion of attractive non-native interactions results in the formation of non-specific encounter complexes that enhance the on-rate of binding, but do not significantly change the binding mechanism. Our study helps explain how being unstructured can confer an advantage in protein target recognition. The simulations are in general agreement with the results of a recently reported nuclear magnetic resonance study, and aid in the interpretation of the experimental binding kinetics.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
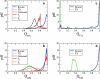
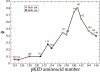
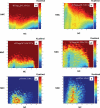
Similar articles
-
Transcriptional activator-coactivator recognition: nascent folding of a kinase-inducible transactivation domain predicts its structure on coactivator binding.Biochemistry. 1998 Apr 28;37(17):5858-66. doi: 10.1021/bi9800808. Biochemistry. 1998. PMID: 9558319
-
Dual role of protein phosphorylation in DNA activator/coactivator binding.Biophys J. 2011 Jan 19;100(2):469-77. doi: 10.1016/j.bpj.2010.11.053. Biophys J. 2011. PMID: 21244843 Free PMC article.
-
Conformational propensities of intrinsically disordered proteins influence the mechanism of binding and folding.Proc Natl Acad Sci U S A. 2015 Aug 4;112(31):9614-9. doi: 10.1073/pnas.1512799112. Epub 2015 Jul 20. Proc Natl Acad Sci U S A. 2015. PMID: 26195786 Free PMC article.
-
The family feud: do proteins with similar structures fold via the same pathway?Curr Opin Struct Biol. 2005 Feb;15(1):42-9. doi: 10.1016/j.sbi.2005.01.011. Curr Opin Struct Biol. 2005. PMID: 15718132 Review.
-
Coupled folding and specific binding: fishing for amphiphilicity.Int J Mol Sci. 2011;12(3):1431-50. doi: 10.3390/ijms12031431. Epub 2011 Feb 24. Int J Mol Sci. 2011. PMID: 21673899 Free PMC article. Review.
Cited by
-
Explicit Characterization of the Free Energy Landscape of pKID-KIX Coupled Folding and Binding.ACS Cent Sci. 2019 Aug 28;5(8):1342-1351. doi: 10.1021/acscentsci.9b00200. Epub 2019 Jul 15. ACS Cent Sci. 2019. PMID: 31482116 Free PMC article.
-
Folding simulations of a de novo designed protein with a betaalphabeta fold.Biophys J. 2010 Jan 20;98(2):321-9. doi: 10.1016/j.bpj.2009.10.018. Biophys J. 2010. PMID: 20338854 Free PMC article.
-
Multiscaled exploration of coupled folding and binding of an intrinsically disordered molecular recognition element in measles virus nucleoprotein.Proc Natl Acad Sci U S A. 2013 Oct 1;110(40):E3743-52. doi: 10.1073/pnas.1308381110. Epub 2013 Sep 16. Proc Natl Acad Sci U S A. 2013. PMID: 24043820 Free PMC article.
-
Nonnative interactions in coupled folding and binding processes of intrinsically disordered proteins.PLoS One. 2010 Nov 4;5(11):e15375. doi: 10.1371/journal.pone.0015375. PLoS One. 2010. PMID: 21079758 Free PMC article.
-
Investigation of sliding DNA clamp dynamics by single-molecule fluorescence, mass spectrometry and structure-based modeling.Nucleic Acids Res. 2018 Apr 6;46(6):3103-3118. doi: 10.1093/nar/gky125. Nucleic Acids Res. 2018. PMID: 29529283 Free PMC article.
References
-
- Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. - PubMed
-
- Minezaki Y, Homma K, Kinjo AR, Nishikawa K. Human transcription factors contain a high fraction of intrinsically disordered regions essential for transcriptional regulation. J Mol Biol. 2006;359:1137–1149. - PubMed
-
- Tompa P. Intrinsically unstructured proteins. Trends Biochem Sci. 2002;27:527–533. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

