Genomic islands in the pathogenic filamentous fungus Aspergillus fumigatus
- PMID: 18404212
- PMCID: PMC2289846
- DOI: 10.1371/journal.pgen.1000046
Genomic islands in the pathogenic filamentous fungus Aspergillus fumigatus
Abstract
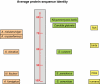
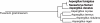
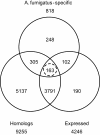
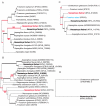
We present the genome sequences of a new clinical isolate of the important human pathogen, Aspergillus fumigatus, A1163, and two closely related but rarely pathogenic species, Neosartorya fischeri NRRL181 and Aspergillus clavatus NRRL1. Comparative genomic analysis of A1163 with the recently sequenced A. fumigatus isolate Af293 has identified core, variable and up to 2% unique genes in each genome. While the core genes are 99.8% identical at the nucleotide level, identity for variable genes can be as low 40%. The most divergent loci appear to contain heterokaryon incompatibility (het) genes associated with fungal programmed cell death such as developmental regulator rosA. Cross-species comparison has revealed that 8.5%, 13.5% and 12.6%, respectively, of A. fumigatus, N. fischeri and A. clavatus genes are species-specific. These genes are significantly smaller in size than core genes, contain fewer exons and exhibit a subtelomeric bias. Most of them cluster together in 13 chromosomal islands, which are enriched for pseudogenes, transposons and other repetitive elements. At least 20% of A. fumigatus-specific genes appear to be functional and involved in carbohydrate and chitin catabolism, transport, detoxification, secondary metabolism and other functions that may facilitate the adaptation to heterogeneous environments such as soil or a mammalian host. Contrary to what was suggested previously, their origin cannot be attributed to horizontal gene transfer (HGT), but instead is likely to involve duplication, diversification and differential gene loss (DDL). The role of duplication in the origin of lineage-specific genes is further underlined by the discovery of genomic islands that seem to function as designated "gene dumps" and, perhaps, simultaneously, as "gene factories".
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

