Beta-globin LCR and intron elements cooperate and direct spatial reorganization for gene therapy
- PMID: 18404216
- PMCID: PMC2271131
- DOI: 10.1371/journal.pgen.1000051
Beta-globin LCR and intron elements cooperate and direct spatial reorganization for gene therapy
Abstract
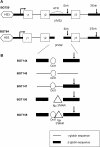
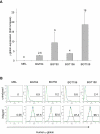
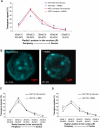
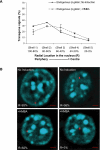
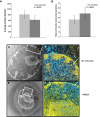
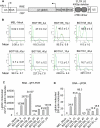
The Locus Control Region (LCR) requires intronic elements within beta-globin transgenes to direct high level expression at all ectopic integration sites. However, these essential intronic elements cannot be transmitted through retrovirus vectors and their deletion may compromise the therapeutic potential for gene therapy. Here, we systematically regenerate functional beta-globin intron 2 elements that rescue LCR activity directed by 5'HS3. Evaluation in transgenic mice demonstrates that an Oct-1 binding site and an enhancer in the intron cooperate to increase expression levels from LCR globin transgenes. Replacement of the intronic AT-rich region with the Igmu 3'MAR rescues LCR activity in single copy transgenic mice. Importantly, a combination of the Oct-1 site, Igmu 3'MAR and intronic enhancer in the BGT158 cassette directs more consistent levels of expression in transgenic mice. By introducing intron-modified transgenes into the same genomic integration site in erythroid cells, we show that BGT158 has the greatest transcriptional induction. 3D DNA FISH establishes that induction stimulates this small 5'HS3 containing transgene and the endogenous locus to spatially reorganize towards more central locations in erythroid nuclei. Electron Spectroscopic Imaging (ESI) of chromatin fibers demonstrates that ultrastructural heterochromatin is primarily perinuclear and does not reorganize. Finally, we transmit intron-modified globin transgenes through insulated self-inactivating (SIN) lentivirus vectors into erythroid cells. We show efficient transfer and robust mRNA and protein expression by the BGT158 vector, and virus titer improvements mediated by the modified intron 2 in the presence of an LCR cassette composed of 5'HS2-4. Our results have important implications for the mechanism of LCR activity at ectopic integration sites. The modified transgenes are the first to transfer intronic elements that potentiate LCR activity and are designed to facilitate correction of hemoglobinopathies using single copy vectors.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
LCR-regulated transgene expression levels depend on the Oct-1 site in the AT-rich region of beta -globin intron-2.Blood. 2003 Feb 15;101(4):1603-10. doi: 10.1182/blood-2002-07-2086. Epub 2002 Sep 26. Blood. 2003. PMID: 12393478
-
Locus control region activity by 5'HS3 requires a functional interaction with beta-globin gene regulatory elements: expression of novel beta/gamma-globin hybrid transgenes.Blood. 2000 May 15;95(10):3242-9. Blood. 2000. PMID: 10807795
-
Evaluation of beta-globin gene therapy constructs in single copy transgenic mice.Nucleic Acids Res. 1997 Mar 15;25(6):1296-302. doi: 10.1093/nar/25.6.1296. Nucleic Acids Res. 1997. PMID: 9092642 Free PMC article.
-
The beta-globin locus control region versus gene therapy vectors: a struggle for expression.Clin Genet. 2001 Jan;59(1):17-24. doi: 10.1034/j.1399-0004.2001.590103.x. Clin Genet. 2001. PMID: 11168020 Review.
-
beta-YAC transgenic mice for studying LCR function.Ann N Y Acad Sci. 1998 Jun 30;850:28-37. doi: 10.1111/j.1749-6632.1998.tb10459.x. Ann N Y Acad Sci. 1998. PMID: 9668524 Review.
Cited by
-
Primordial germ cell-like cells differentiated in vitro from skin-derived stem cells.PLoS One. 2009 Dec 14;4(12):e8263. doi: 10.1371/journal.pone.0008263. PLoS One. 2009. PMID: 20011593 Free PMC article.
-
Isolation of human iPS cells using EOS lentiviral vectors to select for pluripotency.Nat Methods. 2009 May;6(5):370-6. doi: 10.1038/nmeth.1325. Epub 2009 Apr 26. Nat Methods. 2009. PMID: 19404254
-
MECP2 isoform-specific vectors with regulated expression for Rett syndrome gene therapy.PLoS One. 2009 Aug 27;4(8):e6810. doi: 10.1371/journal.pone.0006810. PLoS One. 2009. PMID: 19710912 Free PMC article.
-
Rapid transcriptional pulsing dynamics of high expressing retroviral transgenes in embryonic stem cells.PLoS One. 2012;7(5):e37130. doi: 10.1371/journal.pone.0037130. Epub 2012 May 14. PLoS One. 2012. PMID: 22606340 Free PMC article.
-
Strategies to insulate lentiviral vector-expressed transgenes.Methods Mol Biol. 2010;614:77-100. doi: 10.1007/978-1-60761-533-0_5. Methods Mol Biol. 2010. PMID: 20225037 Free PMC article.
References
-
- Wijgerde M, Grosveld F, Fraser P. Transcription complex stability and chromatin dynamics in vivo. Nature. 1995;377:209–213. - PubMed
-
- Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell. 2002;10:1453–1465. - PubMed
-
- Palstra RJ, Tolhuis B, Splinter E, Nijmeijer R, Grosveld F, et al. The beta-globin nuclear compartment in development and erythroid differentiation. Nat Genet. 2003;35:190–194. - PubMed
-
- Carter D, Chakalova L, Osborne CS, Dai YF, Fraser P. Long-range chromatin regulatory interactions in vivo. Nat Genet. 2002;32:623–626. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

