Protein folding and misfolding: mechanism and principles
- PMID: 18405419
- PMCID: PMC3433742
- DOI: 10.1017/S0033583508004654
Protein folding and misfolding: mechanism and principles
Abstract
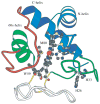
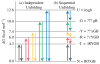
Two fundamentally different views of how proteins fold are now being debated. Do proteins fold through multiple unpredictable routes directed only by the energetically downhill nature of the folding landscape or do they fold through specific intermediates in a defined pathway that systematically puts predetermined pieces of the target native protein into place? It has now become possible to determine the structure of protein folding intermediates, evaluate their equilibrium and kinetic parameters, and establish their pathway relationships. Results obtained for many proteins have serendipitously revealed a new dimension of protein structure. Cooperative structural units of the native protein, called foldons, unfold and refold repeatedly even under native conditions. Much evidence obtained by hydrogen exchange and other methods now indicates that cooperative foldon units and not individual amino acids account for the unit steps in protein folding pathways. The formation of foldons and their ordered pathway assembly systematically puts native-like foldon building blocks into place, guided by a sequential stabilization mechanism in which prior native-like structure templates the formation of incoming foldons with complementary structure. Thus the same propensities and interactions that specify the final native state, encoded in the amino-acid sequence of every protein, determine the pathway for getting there. Experimental observations that have been interpreted differently, in terms of multiple independent pathways, appear to be due to chance misfolding errors that cause different population fractions to block at different pathway points, populate different pathway intermediates, and fold at different rates. This paper summarizes the experimental basis for these three determining principles and their consequences. Cooperative native-like foldon units and the sequential stabilization process together generate predetermined stepwise pathways. Optional misfolding errors are responsible for 3-state and heterogeneous kinetic folding.
Figures




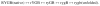
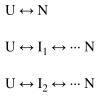
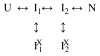
References
-
- Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181:223–230. - PubMed
-
- Bahar I, Wallqvist A, Covell DG, Jernigan RL. Correlation between native-state hydrogen exchange and cooperative residue fluctuations from a simple model. Biochemistry. 1998;37:1067–1075. - PubMed
-
- Bai Y, Englander JJ, Mayne L, Milne JS, Englander SW. Thermodynamic parameters from hydrogen exchange measurements. Methods in Enzymology. 1995a;259:344–356. - PubMed
-
- Bai Y, Englander SW. Future directions in folding: the multi-state nature of protein structure. Proteins: Structure, Function, Genetics. 1996;24:145–151. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

