Negative regulation of quorum-sensing systems in Pseudomonas aeruginosa by ATP-dependent Lon protease
- PMID: 18408026
- PMCID: PMC2446771
- DOI: 10.1128/JB.01873-07
Negative regulation of quorum-sensing systems in Pseudomonas aeruginosa by ATP-dependent Lon protease
Abstract
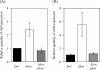
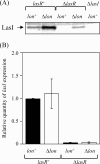
Lon protease, a member of the ATP-dependent protease family, regulates numerous cellular systems by degrading specific substrates. Here, we demonstrate that Lon is involved in the regulation of quorum-sensing (QS) signaling systems in Pseudomonas aeruginosa, an opportunistic human pathogen. The organism has two acyl-homoserine lactone (HSL)-mediated QS systems, LasR/LasI and RhlR/RhlI. Many reports have demonstrated that these two systems are regulated and interconnected by global regulators. We found that lon-disrupted cells overproduce pyocyanin, the biosynthesis of which depends on the RhlR/RhlI system, and show increased levels of a transcriptional regulator, RhlR. The QS systems are organized hierarchically: the RhlR/RhlI system is subordinate to LasR/LasI. To elucidate the mechanism by which Lon negatively regulates RhlR/RhlI, we examined the effect of lon disruption on the LasR/LasI system. We found that Lon represses the expression of LasR/LasI by degrading LasI, an HSL synthase, leading to negative regulation of the RhlR/RhlI system. RhlR/RhlI was also shown to be regulated by Lon independently of LasR/LasI via regulation of RhlI, an HSL synthase. In view of these findings, it is suggested that Lon protease is a powerful negative regulator of both HSL-mediated QS systems in P. aeruginosa.
Figures




Similar articles
-
Evolution of the Pseudomonas aeruginosa quorum-sensing hierarchy.Proc Natl Acad Sci U S A. 2019 Apr 2;116(14):7027-7032. doi: 10.1073/pnas.1819796116. Epub 2019 Mar 8. Proc Natl Acad Sci U S A. 2019. PMID: 30850547 Free PMC article.
-
Analysis of quorum sensing-dependent virulence factor production and its relationship with antimicrobial susceptibility in Pseudomonas aeruginosa respiratory isolates.Clin Microbiol Infect. 2010 Dec;16(12):1770-5. doi: 10.1111/j.1469-0691.2010.03177.x. Clin Microbiol Infect. 2010. PMID: 20132256
-
A rhlI 5' UTR-Derived sRNA Regulates RhlR-Dependent Quorum Sensing in Pseudomonas aeruginosa.mBio. 2019 Oct 8;10(5):e02253-19. doi: 10.1128/mBio.02253-19. mBio. 2019. PMID: 31594819 Free PMC article.
-
An evolving perspective on the Pseudomonas aeruginosa orphan quorum sensing regulator QscR.Front Cell Infect Microbiol. 2014 Oct 28;4:152. doi: 10.3389/fcimb.2014.00152. eCollection 2014. Front Cell Infect Microbiol. 2014. PMID: 25389523 Free PMC article. Review.
-
Pseudomonas aeruginosa quorum sensing modulates immune responses: An updated review article.Immunol Lett. 2017 Oct;190:1-6. doi: 10.1016/j.imlet.2017.07.002. Epub 2017 Jul 8. Immunol Lett. 2017. PMID: 28698104 Review.
Cited by
-
Structural and Molecular Mechanism of CdpR Involved in Quorum-Sensing and Bacterial Virulence in Pseudomonas aeruginosa.PLoS Biol. 2016 Apr 27;14(4):e1002449. doi: 10.1371/journal.pbio.1002449. eCollection 2016 Apr. PLoS Biol. 2016. PMID: 27119725 Free PMC article.
-
Lon Protease Has Multifaceted Biological Functions in Acinetobacter baumannii.J Bacteriol. 2018 Dec 20;201(2):e00536-18. doi: 10.1128/JB.00536-18. Print 2019 Jan 15. J Bacteriol. 2018. PMID: 30348832 Free PMC article.
-
Pseudomonas aeruginosa Lon and ClpXP proteases: roles in linking carbon catabolite repression system with quorum-sensing system.Curr Genet. 2016 Feb;62(1):1-6. doi: 10.1007/s00294-015-0499-5. Epub 2015 Jun 5. Curr Genet. 2016. PMID: 26045103 Review.
-
Developing genome-reduced Pseudomonas chlororaphis strains for the production of secondary metabolites.BMC Genomics. 2017 Sep 11;18(1):715. doi: 10.1186/s12864-017-4127-2. BMC Genomics. 2017. PMID: 28893188 Free PMC article.
-
Evolution of PqsE as a Pseudomonas aeruginosa-specific regulator of LuxR-type receptors: insights from Pseudomonas and Burkholderia.mBio. 2025 May 14;16(5):e0064625. doi: 10.1128/mbio.00646-25. Epub 2025 Apr 8. mBio. 2025. PMID: 40197035 Free PMC article.
References
-
- Becker, A., M. Schmidt, W. Jager, and A. Puhler. 1995. New gentamicin-resistance and lacZ promoter-probe cassettes suitable for insertion mutagenesis and generation of transcriptional fusions. Gene 16237-39. - PubMed
-
- Bretz, J., L. Losada, K. Lisboa, and S. Hutcheson. 2002. Lon protease functions as a negative regulator of type III protein secretion in Pseudomonas syringae. Mol. Microbiol. 45397-409. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

