Investigating early events in receptor binding and translocation of colicin E9 using synchronized cell killing and proteolytic cleavage
- PMID: 18408035
- PMCID: PMC2446772
- DOI: 10.1128/JB.00047-08
Investigating early events in receptor binding and translocation of colicin E9 using synchronized cell killing and proteolytic cleavage
Abstract
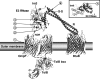
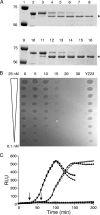
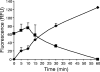
Enzymatic colicins such as colicin E9 (ColE9) bind to BtuB on the cell surface of Escherichia coli and rapidly recruit a second coreceptor, either OmpF or OmpC, through which the N-terminal natively disordered region (NDR) of their translocation domain gains entry into the cell periplasm and interacts with TolB. Previously, we constructed an inactive disulfide-locked mutant ColE9 (ColE9(s-s)) that binds to BtuB and can be reduced with dithiothreitol (DTT) to synchronize cell killing. By introducing unique enterokinase (EK) cleavage sites in ColE9(s-s), we showed that the first 61 residues of the NDR were inaccessible to cleavage when bound to BtuB, whereas an EK cleavage site inserted at residue 82 of the NDR remained accessible. This suggests that most of the NDR is occluded by OmpF shortly after binding to BtuB, whereas the extreme distal region of the NDR is surface exposed before unfolding of the receptor-binding domain occurs. EK cleavage of unique cleavage sites located in the ordered region of the translocation domain or in the distal region of the receptor-binding domain confirmed that these regions of ColE9 remained accessible at the E. coli cell surface. Lack of EK cleavage of the DNase domain of the cell-bound, oxidized ColE9/Im9 complex, and the rapid detection of Alexa Fluor 594-labeled Im9 (Im9(AF)) in the cell supernatant following treatment of cells with DTT, suggested that immunity release occurred immediately after unfolding of the colicin and was not driven by binding to BtuB.
Figures


References
-
- Bonsor, D. A., I. Grishkovskaya, E. J. Dodson, and C. Kleanthous. 2007. Molecular mimicry enables competitive recruitment by a natively disordered protein. J. Am. Chem. Soc. 1294800-4807. - PubMed
-
- Bouveret, E., L. Journet, A. Walburger, E. Cascales, H. Bénédetti, and R. Lloubès. 2002. Analysis of the Escherichia coli Tol-Pal and TonB systems by periplasmic production of Tol, TonB, colicin, or phage capsid soluble domains. Biochimie 84413-421. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

