The beta-strand-loop-beta-strand conformation is marginally populated in beta2-microglobulin (20-41) peptide in solution as revealed by replica exchange molecular dynamics simulations
- PMID: 18408040
- PMCID: PMC2440456
- DOI: 10.1529/biophysj.107.125054
The beta-strand-loop-beta-strand conformation is marginally populated in beta2-microglobulin (20-41) peptide in solution as revealed by replica exchange molecular dynamics simulations
Abstract
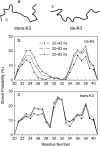
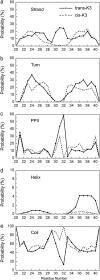
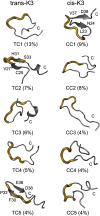
Solid-state NMR study shows that the 22-residue K3 peptide (Ser(20)-Lys(41)) from beta(2)-microglobulin (beta(2)m) adopts a beta-strand-loop-beta-strand conformation in its fibril state. Residue Pro(32) has a trans conformation in the fibril state of the peptide, while it adopts a cis conformation in the native state of full-length beta(2)m. To get insights into the structural properties of the K3 peptide, and determine whether the strand-loop-strand conformation is encoded at the monomeric level, we run all-atom explicit solvent replica exchange molecular dynamics on both the cis and trans variants. Our simulations show that the conformational space of the trans- and cis-K3 peptides is very different, with 1% of the sampled conformations in common at room temperature. In addition, both variants display only 0.3-0.5% of the conformations with beta-strand-loop-beta-strand character. This finding, compared to results on the Alzheimer's Abeta peptide, suggests that the biases toward aggregation leading to the beta-strand-loop-beta-strand conformation in fibrils are peptide-dependent.
Figures



Similar articles
-
Molecular dynamics simulations to gain insights into the stability and morphologies of K3 oligomers from beta2-microglobulin.J Biomol Struct Dyn. 2009 Apr;26(5):549-59. doi: 10.1080/07391102.2009.10507270. J Biomol Struct Dyn. 2009. PMID: 19236105
-
3D structure of amyloid protofilaments of beta2-microglobulin fragment probed by solid-state NMR.Proc Natl Acad Sci U S A. 2006 Nov 28;103(48):18119-24. doi: 10.1073/pnas.0607180103. Epub 2006 Nov 15. Proc Natl Acad Sci U S A. 2006. PMID: 17108084 Free PMC article.
-
Beta2-microglobulin amyloid fragment organization and morphology and its comparison to Abeta suggests that amyloid aggregation pathways are sequence specific.Biochemistry. 2008 Feb 26;47(8):2497-509. doi: 10.1021/bi7019194. Epub 2008 Jan 24. Biochemistry. 2008. PMID: 18215070
-
Limited proteolysis in the investigation of beta2-microglobulin amyloidogenic and fibrillar states.Biochim Biophys Acta. 2005 Nov 10;1753(1):44-50. doi: 10.1016/j.bbapap.2005.09.004. Epub 2005 Sep 23. Biochim Biophys Acta. 2005. PMID: 16213198 Review.
-
Elucidating the Structures of Amyloid Oligomers with Macrocyclic β-Hairpin Peptides: Insights into Alzheimer's Disease and Other Amyloid Diseases.Acc Chem Res. 2018 Mar 20;51(3):706-718. doi: 10.1021/acs.accounts.7b00554. Epub 2018 Mar 6. Acc Chem Res. 2018. PMID: 29508987 Free PMC article. Review.
Cited by
-
Single-molecule visualization determines conformational substate ensembles in β-sheet-rich peptide fibrils.Sci Adv. 2023 Jul 7;9(27):eadg7943. doi: 10.1126/sciadv.adg7943. Epub 2023 Jul 5. Sci Adv. 2023. PMID: 37406110 Free PMC article.
-
K3 fragment of amyloidogenic beta(2)-microglobulin forms ion channels: implication for dialysis related amyloidosis.J Am Chem Soc. 2009 Oct 21;131(41):14938-45. doi: 10.1021/ja9049299. J Am Chem Soc. 2009. PMID: 19824733 Free PMC article.
-
Amyloid β Protein and Alzheimer's Disease: When Computer Simulations Complement Experimental Studies.Chem Rev. 2015 May 13;115(9):3518-63. doi: 10.1021/cr500638n. Epub 2015 Mar 19. Chem Rev. 2015. PMID: 25789869 Free PMC article. Review.
-
Protein Ensembles: How Does Nature Harness Thermodynamic Fluctuations for Life? The Diverse Functional Roles of Conformational Ensembles in the Cell.Chem Rev. 2016 Jun 8;116(11):6516-51. doi: 10.1021/acs.chemrev.5b00562. Epub 2016 Jan 25. Chem Rev. 2016. PMID: 26807783 Free PMC article. Review.
-
The Early Phase of β2-Microglobulin Aggregation: Perspectives From Molecular Simulations.Front Mol Biosci. 2020 Sep 29;7:578433. doi: 10.3389/fmolb.2020.578433. eCollection 2020. Front Mol Biosci. 2020. PMID: 33134317 Free PMC article. Review.
References
-
- Lashuel, H. A., and P. T. J. Lansbury. 2006. Are amyloid diseases caused by protein aggregates that mimic bacterial pore-forming toxins? Q. Rev. Biophys. 36:167–201. - PubMed
-
- Kayed, R., E. Head, J. L. Thompson, T. M. McIntire, S. C. Milton, C. W. Cotman, and C. G. Glabe. 2003. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 300:486–489. - PubMed
-
- Kayed, R., and C. G. Glabe. 2006. Conformation-dependent anti-amyloid oligomer antibodies. Methods Enzymol. 413:326–344. - PubMed
-
- Chiba, T., T. Hagihara, Y. Higurashi, K. Hasegawa, H. Naiki, and Y. Goto. 2003. Amyloid fibril formation in the context of full-length protein: effects of proline mutations on the amyloid fibril formation of β2-microglobulin. J. Biol. Chem. 278:47016–47024. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

