The genome of Bacillus coahuilensis reveals adaptations essential for survival in the relic of an ancient marine environment
- PMID: 18408155
- PMCID: PMC2311347
- DOI: 10.1073/pnas.0800981105
The genome of Bacillus coahuilensis reveals adaptations essential for survival in the relic of an ancient marine environment
Abstract
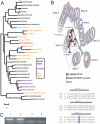
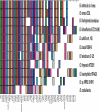
The Cuatro Ciénegas Basin (CCB) in the central part of the Chihuahan desert (Coahuila, Mexico) hosts a wide diversity of microorganisms contained within springs thought to be geomorphological relics of an ancient sea. A major question remaining to be answered is whether bacteria from CCB are ancient marine bacteria that adapted to an oligotrophic system poor in NaCl, rich in sulfates, and with extremely low phosphorus levels (<0.3 microM). Here, we report the complete genome sequence of Bacillus coahuilensis, a sporulating bacterium isolated from the water column of a desiccation lagoon in CCB. At 3.35 Megabases this is the smallest genome sequenced to date of a Bacillus species and provides insights into the origin, evolution, and adaptation of B. coahuilensis to the CCB environment. We propose that the size and complexity of the B. coahuilensis genome reflects the adaptation of an ancient marine bacterium to a novel environment, providing support to a "marine isolation origin hypothesis" that is consistent with the geology of CCB. This genomic adaptation includes the acquisition through horizontal gene transfer of genes involved in phosphorous utilization efficiency and adaptation to high-light environments. The B. coahuilensis genome sequence also revealed important ecological features of the bacterial community in CCB and offers opportunities for a unique glimpse of a microbe-dominated world last seen in the Precambrian.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Minckley WL. Environments of the Bolson of Cuatro Ciénegas, Coahuila, México, with Special Reference to the Aquatic Biota. El Paso, TX: Texas Western Press; 1969. University of Texas: El Paso Science Series.
-
- Elser JJ, et al. Effects of phosphorus enrichment and grazing snails on modern stromatolitic microbial communities. Freshwater Biol. 2005;50:1808–1825.
-
- Cerritos R, et al. Bacillus coahuilensis sp. nov. a new moderately halophilic species from different pozas in the Cuatro Ciénegas Valley in Coahuila, México. Int J Syst Evol Microbiol. 2008;58:919–923. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

