An overlapping essential gene in the Potyviridae
- PMID: 18408156
- PMCID: PMC2311343
- DOI: 10.1073/pnas.0800468105
An overlapping essential gene in the Potyviridae
Abstract
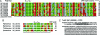
The family Potyviridae includes >30% of known plant virus species, many of which are of great agricultural significance. These viruses have a positive sense RNA genome that is approximately 10 kb long and contains a single long ORF. The ORF is translated into a large polyprotein, which is cleaved into approximately 10 mature proteins. We report the discovery of a short ORF embedded within the P3 cistron of the polyprotein but translated in the +2 reading-frame. The ORF, termed pipo, is conserved and has a strong bioinformatic coding signature throughout the large and diverse Potyviridae family. Mutations that knock out expression of the PIPO protein in Turnip mosaic potyvirus but leave the polyprotein amino acid sequence unaltered are lethal to the virus. Immunoblotting with antisera raised against two nonoverlapping 14-aa antigens, derived from the PIPO amino acid sequence, reveals the expression of an approximately 25-kDa PIPO fusion product in planta. This is consistent with expression of PIPO as a P3-PIPO fusion product via ribosomal frameshifting or transcriptional slippage at a highly conserved G(1-2)A(6-7) motif at the 5' end of pipo. This discovery suggests that other short overlapping genes may remain hidden even in well studied virus genomes (as well as cellular organisms) and demonstrates the utility of the software package MLOGD as a tool for identifying such genes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
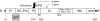




References
-
- Firth AE, Brown CM. Detecting overlapping coding sequences with pairwise alignments. Bioinformatics. 2005;21:282–292. - PubMed
-
- Riechmann JL, Laín S, García JA. Highlights and prospects of potyvirus molecular biology. J Gen Virol. 1992;73:1–16. - PubMed
-
- Berger PH, et al. In: Virus Taxonomy: Eighth Report of the International Committee on the Taxonomy of Viruses. Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. San Diego: Elsevier Academic; 2005. pp. 819–841.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

