Mitotic phosphorylation stimulates DNA relaxation activity of human topoisomerase I
- PMID: 18408216
- PMCID: PMC2423254
- DOI: 10.1074/jbc.M802246200
Mitotic phosphorylation stimulates DNA relaxation activity of human topoisomerase I
Abstract
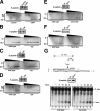
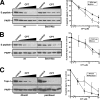
Human DNA topoisomerase I (topo I) is an essential mammalian enzyme that regulates DNA supercoiling during transcription and replication. In addition, topo I is specifically targeted by the anticancer compound camptothecin and its derivatives. Previous studies have indicated that topo I is a phosphoprotein and that phosphorylation stimulates its DNA relaxation activity. The locations of most topo I phosphorylation sites have not been identified, preventing a more detailed examination of this modification. To address this issue, mass spectrometry was used to identify four topo I residues that are phosphorylated in intact cells: Ser(10), Ser(21), Ser(112), and Ser(394). Immunoblotting using anti-phosphoepitope antibodies demonstrated that these sites are phosphorylated during mitosis. In vitro kinase assays demonstrated that Ser(10) can be phosphorylated by casein kinase II, Ser(21) can be phosphorylated by protein kinase Calpha, and Ser(112) and Ser(394) can be phosphorylated by Cdk1. When wild type topo I was pulled down from mitotic cells and dephosphorylated with alkaline phosphatase, topo I activity decreased 2-fold. Likewise, topo I polypeptide with all four phosphorylation sites mutated to alanine exhibited 2-fold lower DNA relaxation activity than wild type topo I after isolation from mitotic cells. Further mutational analysis demonstrated that Ser(21) phosphorylation was responsible for this change. Consistent with these results, wild type topo I (but not S21A topo I) exhibited increased sensitivity to camptothecin-induced trapping on DNA during mitosis. Collectively these results indicate that topo I is phosphorylated during mitosis at multiple sites, one of which enhances DNA relaxation activity in vitro and interaction with DNA in cells.
Figures
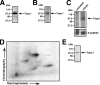

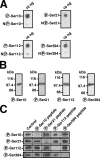
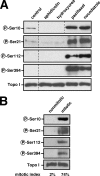

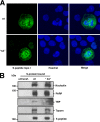
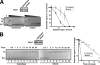
References
-
- Wang, J. C. (2002) Nat. Rev. Mol. Cell Biol. 3 430-440 - PubMed
-
- Champoux, J. J. (2001) Annu. Rev. Biochem. 70 369-413 - PubMed
-
- Madden, K. R., and Champoux, J. J. (1992) Cancer Res. 52 525-532 - PubMed
-
- Koster, D. A., Palle, K., Bot, E. S., Bjornsti, M. A., and Dekker, N. H. (2007) Nature 448 213-217 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

