Smooth skyride through a rough skyline: Bayesian coalescent-based inference of population dynamics
- PMID: 18408232
- PMCID: PMC3302198
- DOI: 10.1093/molbev/msn090
Smooth skyride through a rough skyline: Bayesian coalescent-based inference of population dynamics
Abstract
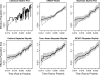
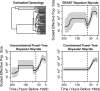
Kingman's coalescent process opens the door for estimation of population genetics model parameters from molecular sequences. One paramount parameter of interest is the effective population size. Temporal variation of this quantity characterizes the demographic history of a population. Because researchers are rarely able to choose a priori a deterministic model describing effective population size dynamics for data at hand, nonparametric curve-fitting methods based on multiple change-point (MCP) models have been developed. We propose an alternative to change-point modeling that exploits Gaussian Markov random fields to achieve temporal smoothing of the effective population size in a Bayesian framework. The main advantage of our approach is that, in contrast to MCP models, the explicit temporal smoothing does not require strong prior decisions. To approximate the posterior distribution of the population dynamics, we use efficient, fast mixing Markov chain Monte Carlo algorithms designed for highly structured Gaussian models. In a simulation study, we demonstrate that the proposed temporal smoothing method, named Bayesian skyride, successfully recovers "true" population size trajectories in all simulation scenarios and competes well with the MCP approaches without evoking strong prior assumptions. We apply our Bayesian skyride method to 2 real data sets. We analyze sequences of hepatitis C virus contemporaneously sampled in Egypt, reproducing all key known aspects of the viral population dynamics. Next, we estimate the demographic histories of human influenza A hemagglutinin sequences, serially sampled throughout 3 flu seasons.
Figures





References
-
- Bernardinelli L, Clayton D, Montomoli C. Bayesian estimates of disease maps: how important are priors? Stat Med. 1995;14:2411–2431. - PubMed
-
- Biek R, Drummond A, Poss M. A virus reveals population structure and recent demographic history of its carnivore host. Science. 2006;311:538–541. - PubMed
-
- Chib S, Jeliazkov I. Marginal likelihood from the Metropolis-Hastings output. J Am Stat Assoc. 2001;96:270–281.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

