Nucleosome organization in the Drosophila genome
- PMID: 18408708
- PMCID: PMC2735122
- DOI: 10.1038/nature06929
Nucleosome organization in the Drosophila genome
Abstract
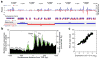
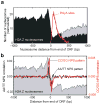
Comparative genomics of nucleosome positions provides a powerful means for understanding how the organization of chromatin and the transcription machinery co-evolve. Here we produce a high-resolution reference map of H2A.Z and bulk nucleosome locations across the genome of the fly Drosophila melanogaster and compare it to that from the yeast Saccharomyces cerevisiae. Like Saccharomyces, Drosophila nucleosomes are organized around active transcription start sites in a canonical -1, nucleosome-free region, +1 arrangement. However, Drosophila does not incorporate H2A.Z into the -1 nucleosome and does not bury its transcriptional start site in the +1 nucleosome. At thousands of genes, RNA polymerase II engages the +1 nucleosome and pauses. How the transcription initiation machinery contends with the +1 nucleosome seems to be fundamentally different across major eukaryotic lines.
Figures



References
-
- Albert I, et al. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. - PubMed
-
- Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
-
- Lee W, et al. A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet. 2007;39:1235–1244. - PubMed
-
- Mito Y, Henikoff JG, Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nat Genet. 2005;37:1090–1097. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

