ATP-dependent chromatin remodeling shapes the DNA replication landscape
- PMID: 18408730
- PMCID: PMC2678716
- DOI: 10.1038/nsmb.1419
ATP-dependent chromatin remodeling shapes the DNA replication landscape
Abstract
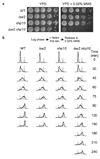
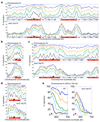
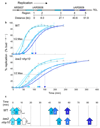
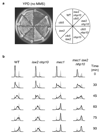
The eukaryotic DNA replication machinery must traverse every nucleosome in the genome during S phase. As nucleosomes are generally inhibitory to DNA-dependent processes, chromatin structure must undergo extensive reorganization to facilitate DNA synthesis. However, the identity of chromatin-remodeling factors involved in replication and how they affect DNA synthesis is largely unknown. Here we show that two highly conserved ATP-dependent chromatin-remodeling complexes in Saccharomyces cerevisiae, Isw2 and Ino80, function in parallel to promote replication fork progression. As a result, Isw2 and Ino80 have especially important roles for replication of late-replicating regions during periods of replication stress. Both Isw2 and Ino80 complexes are enriched at sites of replication, suggesting that these complexes act directly to promote fork progression. These findings identify ATP-dependent chromatin-remodeling complexes that promote DNA replication and define a specific stage of replication that requires remodeling for normal function.
Figures

References
-
- Becker PB, Horz W. ATP-dependent nucleosome remodeling. Annu Rev Biochem. 2002;71:247–273. - PubMed
-
- Smith CL, Peterson CL. ATP-dependent chromatin remodeling. Curr Top Dev Biol. 2005;65:115–148. - PubMed
-
- Narlikar GJ, Fan HY, Kingston RE. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–487. - PubMed
-
- Varga-Weisz P. Chromatin remodeling factors and DNA replication. Prog Mol Subcell Biol. 2005;38:1–30. - PubMed
-
- Ataian Y, Krebs JE. Five repair pathways in one context: chromatin modification during DNA repair. Biochem Cell Biol. 2006;84:490–504. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

