Inactivation of a human kinetochore by specific targeting of chromatin modifiers
- PMID: 18410728
- PMCID: PMC2311382
- DOI: 10.1016/j.devcel.2008.02.001
Inactivation of a human kinetochore by specific targeting of chromatin modifiers
Abstract
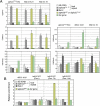
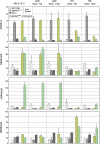
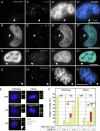
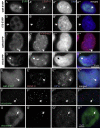
We have used a human artificial chromosome (HAC) to manipulate the epigenetic state of chromatin within an active kinetochore. The HAC has a dimeric alpha-satellite repeat containing one natural monomer with a CENP-B binding site, and one completely artificial synthetic monomer with the CENP-B box replaced by a tetracycline operator (tetO). This HAC exhibits normal kinetochore protein composition and mitotic stability. Targeting of several tet-repressor (tetR) fusions into the centromere had no effect on kinetochore function. However, altering the chromatin state to a more open configuration with the tTA transcriptional activator or to a more closed state with the tTS transcription silencer caused missegregation and loss of the HAC. tTS binding caused the loss of CENP-A, CENP-B, CENP-C, and H3K4me2 from the centromere accompanied by an accumulation of histone H3K9me3. Our results reveal that a dynamic balance between centromeric chromatin and heterochromatin is essential for vertebrate kinetochore activity.
Figures




References
-
- Agata Y., Matsuda E., Shimizu A. Two novel Kruppel-associated box-containing zinc-finger proteins, KRAZ1 and KRAZ2, repress transcription through functional interaction with the corepressor KAP-1 (TIF1β/KRIP-1) J. Biol. Chem. 1999;274:16412–16422. - PubMed
-
- Allshire R.C., Javerzat J.-P., Redhead N.J., Cranston G. Position effect variegation at fission yeast centromeres. Cell. 1994;76:157–169. - PubMed
-
- Alonso A., Mahmood R., Li S., Cheung F., Yoda K., Warburton P.E. Genomic microarray analysis reveals distinct locations for the CENP-A binding domains in three human chromosome 13q32 neocentromeres. Hum. Mol. Genet. 2003;12:2711–2721. - PubMed
-
- Black B.E., Foltz D.R., Chakravarthy S., Luger K., Woods V.L., Jr., Cleveland D.W. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

