Genome level analysis of rice mRNA 3'-end processing signals and alternative polyadenylation
- PMID: 18411206
- PMCID: PMC2396415
- DOI: 10.1093/nar/gkn158
Genome level analysis of rice mRNA 3'-end processing signals and alternative polyadenylation
Abstract
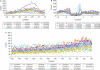
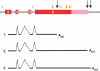
The position of a poly(A) site of eukaryotic mRNA is determined by sequence signals in pre-mRNA and a group of polyadenylation factors. To reveal rice poly(A) signals at a genome level, we constructed a dataset of 55 742 authenticated poly(A) sites and characterized the poly(A) signals. This resulted in identifying the typical tripartite cis-elements, including FUE, NUE and CE, as previously observed in Arabidopsis. The average size of the 3'-UTR was 289 nucleotides. When mapped to the genome, however, 15% of these poly(A) sites were found to be located in the currently annotated intergenic regions. Moreover, an extensive alternative polyadenylation profile was evident where 50% of the genes analyzed had more than one unique poly(A) site (excluding microheterogeneity sites), and 13% had four or more poly(A) sites. About 4% of the analyzed genes possessed alternative poly(A) sites at their introns, 5'-UTRs, or protein coding regions. The authenticity of these alternative poly(A) sites was partially confirmed using MPSS data. Analysis of nucleotide profile and signal patterns indicated that there may be a different set of poly(A) signals for those poly(A) sites found in the coding regions. Based on the features of rice poly(A) signals, an updated algorithm termed PASS-Rice was designed to predict poly(A) sites.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

