The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2
- PMID: 18411277
- PMCID: PMC3258899
- DOI: 10.1074/jbc.C800074200
The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2
Abstract
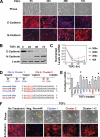
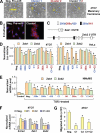
MicroRNAs are small non-coding RNA molecules that can regulate gene expression by interacting with multiple mRNAs and inducing either translation suppression or degradation of mRNA. Recently, several miRNAs were identified as either promoters or suppressors of metastasis. However, it is unclear in which step(s) of the multistep metastatic cascade these miRNAs play a defined functional role. To study the functional importance of miRNAs in epithelial-mesenchymal transition (EMT), a process thought to initiate metastasis by enhancing the motility of tumor cells, we used a well established in vitro EMT assay: transforming growth factor-beta-induced EMT in NMuMG murine mammary epithelial cells. We found that members of the miR-200 family, organized as two clusters in the genome, were repressed during EMT. Overexpression of each miRNA individually or as clusters in NMuMG cells hindered EMT by enhancing E-cadherin expression through direct targeting of ZEB1 and ZEB2, which encode transcriptional repressors of E-cadherin. In the 4TO7 mouse carcinoma cell line, which expresses low levels of endogenous E-cadherin and displays a mesenchymal phenotype, ectopic expression of the miR-200 family miRNAs significantly increased E-cadherin expression and altered cell morphology to an epithelial phenotype. Furthermore, ectopic expression of each miR-200 miRNA cluster significantly reduced the in vitro motility of 4TO7 cells in migration assays. These results suggested that loss of expression of the miR-200 family members may play a critical role in the repression of E-cadherin by ZEB1 and ZEB2 during EMT, thereby enhancing migration and invasion during cancer progression.
Figures
References
-
- Lee, Y., Ahn, C., Han, J., Choi, H., Kim, J., Yim, J., Lee, J., Provost, P., Radmark, O., Kim, S., and Kim, V. N. (2003) Nature 425 415–419 - PubMed
-
- Hutvagner, G., McLachlan, J., Pasquinelli, A. E., Balint, E., Tuschl, T., and Zamore, P. D. (2001) Science 293 834–838 - PubMed
-
- Wienholds, E., Kloosterman, W. P., Miska, E., Alvarez-Saavedra, E., Berezikov, E., de Bruijn, E., Horvitz, H. R., Kauppinen, S., and Plasterk, R. H. (2005) Science 309 310–311 - PubMed
-
- Yi, R., O'Carroll, D., Pasolli, H. A., Zhang, Z., Dietrich, F. S., Tarakhovsky, A., and Fuchs, E. (2006) Nat. Genet. 38 356–362 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

