The surface-exposed carboxyl region of Mycoplasma pneumoniae elongation factor Tu interacts with fibronectin
- PMID: 18411296
- PMCID: PMC2446705
- DOI: 10.1128/IAI.00173-08
The surface-exposed carboxyl region of Mycoplasma pneumoniae elongation factor Tu interacts with fibronectin
Abstract
Mycoplasma pneumoniae is a bacterial pathogen of the human respiratory tract that causes a wide range of airway diseases as well as extrapulmonary symptoms. It possesses a distinct, differentiated terminal structure, termed the attachment organelle, that mediates adherence to the host respiratory epithelium. Previously, we reported that surface-associated M. pneumoniae elongation factor Tu (EF-Tu, also called MPN665) serves as a fibronectin (Fn)-binding protein, facilitating interactions between mycoplasmas and extracellular matrix. In the present study, we determined that binding of M. pneumoniae EF-Tu to Fn is primarily mediated by the EF-Tu carboxyl region. A 179-amino-acid region spanning the carboxyl terminus (designated EC; amino acids 192 to 394) binds Fn in a dose-dependent manner. Further analysis of carboxyl constructs (ED3 and ED4) and their deletion truncations (ED3.1, ED3.2, and ED4.1) revealed that the carboxyl region possessed two distinct sites with different Fn-binding efficiencies. Immunogold electron microscopy using antibodies raised against recombinant ED3 and ED4 demonstrated the surface accessibility of the EF-Tu carboxyl region. Competitive binding assays using intact radiolabeled mycoplasmas and purified recombinant ED3 and ED4 proteins, along with antibody blocking assays, reinforced the role of the surface-exposed EF-Tu carboxyl region in Fn binding. Alkali and high-salt treatment of mycoplasma membranes and Triton X-114-partitioned mycoplasma fractions confirmed the stable association of EF-Tu within the mycoplasma membrane. These observations highlight the unique, multifaceted, and unpredictable role of the classically defined cytoplasmic protein EF-Tu relative to cellular function, compartmentalization, and topography.
Figures

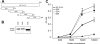
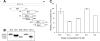



References
-
- Alvarez, R. A., M. W. Blaylock, and J. B. Baseman. 2003. Surface localized glyceraldehyde-3-phosphate dehydrogenase of Mycoplasma genitalium binds mucin. Mol. Microbiol. 481417-1425. - PubMed
-
- Bergmann, S., M. Rohde, G. S. Chhatwal, and S. Hammerschmidt. 2001. Alpha-Enolase of Streptococcus pneumoniae is a plasmin(ogen)-binding protein displayed on the bacterial cell surface. Mol. Microbiol. 401273-1287. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

