Multiscale modeling of nucleic acids: insights into DNA flexibility
- PMID: 18412139
- PMCID: PMC2561918
- DOI: 10.1002/bip.21000
Multiscale modeling of nucleic acids: insights into DNA flexibility
Abstract
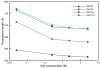
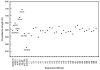
The elastic rod theory is used together with all-atom normal mode analysis in implicit solvent to characterize the mechanical flexibility of duplex DNA. The bending, twisting, stretching rigidities extracted from all-atom simulations (on linear duplexes from 60 to 150 base pairs in length and from 94-bp minicircles) are in reasonable agreement with experimental results. We focus on salt concentration and sequence effects on the overall flexibility. Bending persistence lengths are about 20% higher than most experimental estimates, but the transition from low-salt to high-salt behavior is reproduced well, as is the dependence of the stretching modulus on salt (which is opposite to that of bending). CTG and CGG trinucleotide repeats, responsible for several degenerative disorders, are found to be more flexible than random DNA, in agreement with several recent studies, whereas poly(dA).poly(dT) is the stiffest sequence we have encountered. The results suggest that current all-atom potentials, which were parameterized on small molecules and short oligonucleotides, also provide a useful description of duplex DNA at much longer length scales.
Figures










References
-
- Cramer CJ, Truhlar DG. Implicit solvation models: Equilibria, structure, spectra, and dynamics. Chem Rev. 1999;99:2161–2200. - PubMed
-
- Feig M, Brooks CL. Recent advances in the development and application of implicit solvent models in biomolecule simulations. Curr Opin Struct Biol. 2004;14:217–224. - PubMed
-
- Bashford D, Case DA. Generalized born models of macromolecular solvation effects. Annu Rev Phys Chem. 2000;51:129–152. - PubMed
-
- Brown RA, Case DA. Second derivatives in generalized born theory. J Comput Chem. 2006;27:1662–1675. - PubMed
-
- Tsui V, Case DA. Theory and applications of the generalized born solvation model in macro-molecular simulations. Biopolymers. 2000;56:275–291. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

