Global regulatory logic for specification of an embryonic cell lineage
- PMID: 18413610
- PMCID: PMC2329687
- DOI: 10.1073/pnas.0711220105
Global regulatory logic for specification of an embryonic cell lineage
Abstract
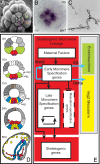
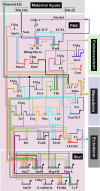
Explanation of a process of development must ultimately be couched in the terms of the genomic regulatory code. Specification of an embryonic cell lineage is driven by a network of interactions among genes encoding transcription factors. Here, we present the gene regulatory network (GRN) that directs the specification of the skeletogenic micromere lineage of the sea urchin embryo. The GRN now includes all regulatory genes expressed in this lineage up to late blastula stage, as identified in a genomewide survey. The architecture of the GRN was established by a large-scale perturbation analysis in which the expression of each gene in the GRN was cut off by use of morpholinos, and the effects on all other genes were measured quantitatively. Several cis-regulatory analyses provided additional evidence. The explanatory power of the GRN suffices to provide a causal explanation for all observable developmental functions of the micromere lineage during the specification period. These functions are: (i) initial acquisition of identity through transcriptional interpretation of localized maternal cues; (ii) activation of specific regulatory genes by use of a double negative gate; (iii) dynamic stabilization of the regulatory state by activation of a feedback subcircuit; (iv) exclusion of alternative regulatory states; (v) presentation of a signal required by the micromeres themselves and of two different signals required for development of adjacent endomesodermal lineages; and (vi) lineage-specific activation of batteries of skeletogenic genes. The GRN precisely predicts gene expression responses and provides a coherent explanation of the biology of specification.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Comment in
-
Gene regulatory networks and embryonic specification.Proc Natl Acad Sci U S A. 2008 Apr 22;105(16):5951-2. doi: 10.1073/pnas.0801434105. Epub 2008 Apr 15. Proc Natl Acad Sci U S A. 2008. PMID: 18417452 Free PMC article. No abstract available.
References
-
- Oliveri P, Davidson EH. Gene regulatory network controlling embryonic specification in the sea urchin. Curr Opin Genet Dev. 2004;14:351–360. - PubMed
-
- Davidson EH. The Regulatory Genome: Gene Regulatory Networks in Development and Evolution. San Diego: Academic; 2006.
-
- Davidson EH, et al. A genomic regulatory network for development. Science. 2002;295:1669–1678. - PubMed
-
- Howard-Ashby M, et al. Identification and characterization of homeobox transcription factor genes in Strongylocentrotus purpuratus and their expression in embryonic development. Dev Biol. 2006;300:74–89. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

