Alteration of cyclin D1 transcript elongation by a mutated transcription factor up-regulates the oncogenic D1b splice isoform in cancer
- PMID: 18413612
- PMCID: PMC2329709
- DOI: 10.1073/pnas.0710748105
Alteration of cyclin D1 transcript elongation by a mutated transcription factor up-regulates the oncogenic D1b splice isoform in cancer
Abstract
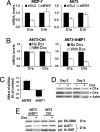
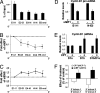
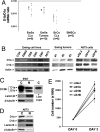
Pre-mRNA splicing and polyadenylation are tightly connected to transcription, and transcriptional stimuli and elongation dynamics can affect mRNA maturation. However, whether this regulatory mechanism has a physio/pathological impact is not known. In cancer, where splice variant expression is often deregulated, many mutated oncogenes are transcriptional regulators. In particular, the Ewing sarcoma (EwSa) oncogene, resulting from a fusion of the EWS and FLI1 genes, encodes a well characterized transcription factor. EWS-FLI1 directly stimulates transcription of the CCND1 protooncogene encoding cyclin D1a and a less abundant but more oncogenic splice isoform, D1b. We show that, although both EWS and EWS-FLI1 enhance cyclin D1 gene expression, they regulate the D1b/D1a transcript ratio in an opposite manner. Detailed analyses of RNA polymerase dynamics along the gene and of the effects of an inhibitor of elongation show that EWS-FLI1 favors D1b isoform expression by decreasing the elongation rate, whereas EWS has opposite effects. As a result, the D1b/D1a ratio is elevated in EwSa cell lines and tumors. The endogenous D1b protein is enriched in nuclei, where the oncogenic activity of cyclin D1 is known to occur, and depleting D1b in addition to D1a results in a stronger reduction of EwSa cell growth than depleting D1a only. These data show that elevated expression of a splice isoform in cancer can be due to an alteration of the transcription process by a mutated transcriptional regulator and provide evidence for a physio/pathological impact of the coupling between transcription and mRNA maturation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
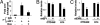
Similar articles
-
Coupled alteration of transcription and splicing by a single oncogene: boosting the effect on cyclin D1 activity.Cell Cycle. 2008 Aug;7(15):2299-305. doi: 10.4161/cc.6445. Epub 2008 Jun 17. Cell Cycle. 2008. PMID: 18677114 Review.
-
Oncogenic fusion protein EWS-FLI1 is a network hub that regulates alternative splicing.Proc Natl Acad Sci U S A. 2015 Mar 17;112(11):E1307-16. doi: 10.1073/pnas.1500536112. Epub 2015 Mar 3. Proc Natl Acad Sci U S A. 2015. PMID: 25737553 Free PMC article.
-
The Promoter-Associated Noncoding RNA pncCCND1_B Assembles a Protein-RNA Complex to Regulate Cyclin D1 Transcription in Ewing Sarcoma.Cancer Res. 2019 Jul 15;79(14):3570-3582. doi: 10.1158/0008-5472.CAN-18-2403. Epub 2019 May 9. Cancer Res. 2019. PMID: 31072811
-
PI3K/AKT signaling modulates transcriptional expression of EWS/FLI1 through specificity protein 1.Oncotarget. 2015 Oct 6;6(30):28895-910. doi: 10.18632/oncotarget.5000. Oncotarget. 2015. PMID: 26336820 Free PMC article.
-
EWS-FLI1 in Ewing's sarcoma: real targets and collateral damage.Adv Exp Med Biol. 2006;587:41-52. doi: 10.1007/978-1-4020-5133-3_4. Adv Exp Med Biol. 2006. PMID: 17163154 Review.
Cited by
-
Impact of G1 phase kinetics on the acquisition of stemness in cancer cells: the critical role of cyclin D.Mol Biol Rep. 2025 Feb 14;52(1):230. doi: 10.1007/s11033-025-10351-3. Mol Biol Rep. 2025. PMID: 39951181 Review.
-
Ewing sarcoma fusion protein EWSR1/FLI1 interacts with EWSR1 leading to mitotic defects in zebrafish embryos and human cell lines.Cancer Res. 2009 May 15;69(10):4363-71. doi: 10.1158/0008-5472.CAN-08-3229. Epub 2009 May 5. Cancer Res. 2009. PMID: 19417137 Free PMC article.
-
Genotoxic stress inhibits Ewing sarcoma cell growth by modulating alternative pre-mRNA processing of the RNA helicase DHX9.Oncotarget. 2015 Oct 13;6(31):31740-57. doi: 10.18632/oncotarget.5033. Oncotarget. 2015. PMID: 26450900 Free PMC article.
-
Influence of CCND1 G870A polymorphism on the risk of HBV-related HCC and cyclin D1 splicing variant expression in Chinese population.Tumour Biol. 2015 Sep;36(9):6891-900. doi: 10.1007/s13277-015-3401-7. Epub 2015 Apr 8. Tumour Biol. 2015. PMID: 25851350 Free PMC article.
-
EWSR1 binds the hepatitis C virus cis-acting replication element and is required for efficient viral replication.J Virol. 2013 Jun;87(12):6625-34. doi: 10.1128/JVI.01006-12. Epub 2013 Apr 3. J Virol. 2013. PMID: 23552423 Free PMC article.
References
-
- Stamm S, et al. Function of alternative splicing. Gene. 2005;344:1–20. - PubMed
-
- Srebrow A, Kornblihtt AR. The connection between splicing and cancer. J Cell Sci. 2006;119:2635–2641. - PubMed
-
- Venables JP. Unbalanced alternative splicing and its significance in cancer. BioEssays. 2006;28:378–386. - PubMed
-
- Caceres JF, Kornblihtt AR. Alternative splicing: Multiple control mechanisms and involvement in human disease. Trends Genet. 2002;18:186–193. - PubMed
-
- Bentley DL. Rules of engagement: Co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol. 2005;17:251–256. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

