Synthetic gene brushes: a structure-function relationship
- PMID: 18414482
- PMCID: PMC2387232
- DOI: 10.1038/msb.2008.20
Synthetic gene brushes: a structure-function relationship
Abstract
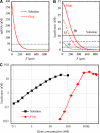
We present the assembly of gene brushes by means of a photolithographic approach that allows us to control the density of end-immobilized linear double-stranded DNA polymers coding for entire genes. For 2 kbp DNAs, the mean distance varies from 300 nm, where DNAs are dilute and assume relaxed conformations, down to 30 nm, where steric repulsion at dense packing forces stretching out. We investigated the gene-to-protein relationship of firefly luciferase under the T7/E.Coli-extract expression system, as well as transcription-only reactions with T7 RNA polymerase, and found both systems to be highly sensitive to brush density, conformation, and orientation. A 'structure-function' picture emerges in which extension of genes induced by moderate packing exposes coding sequences and improves their interaction with the transcription/translation machinery. However, tighter packing impairs the penetration of the machinery into the brush. The response of expression to two-dimensional gene crowding at the nanoscale identifies gene brushes as basic controllable units en route to multicomponent synthetic systems. In turn, these brushes could deepen our understanding of biochemical reactions taking place under confinement and molecular crowding in living cells.
Figures


References
-
- Buxboim A, Bar-Dagan M, Frydman V, Zbaida D, Morpurgo M, Bar-Ziv R (2007) A single-step photolithographic interface for cell-free gene expression and active biochips. Small 3: 500–510 - PubMed
-
- Daube SS, Arad T, Bar-Ziv R (2007) Cell-free co-synthesis of protein nanoassemblies: tubes, rings, and doughnuts. Nano Lett 7: 638–641 - PubMed
-
- Dittmer WU, Kempter S, Radler JO, Simmel FC (2005) Using gene regulation to program DNA-based molecular devices. Small 1: 709–712 - PubMed
-
- Dittmer WU, Simmel FC (2004) Transcriptional control of DNA-based nanomachines. Nano Lett 4: 689–691
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

