Analyses of in vivo interaction and mobility of two spliceosomal proteins using FRAP and BiFC
- PMID: 18414657
- PMCID: PMC2278372
- DOI: 10.1371/journal.pone.0001953
Analyses of in vivo interaction and mobility of two spliceosomal proteins using FRAP and BiFC
Abstract
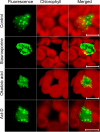
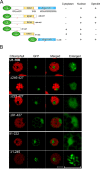
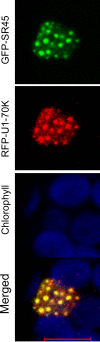
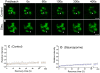
U1-70K, a U1 snRNP-specific protein, and serine/arginine-rich (SR) proteins are components of the spliceosome and play critical roles in both constitutive and alternative pre-mRNA splicing. However, the mobility properties of U1-70K, its in vivo interaction with SR proteins, and the mobility of the U1-70K-SR protein complex have not been studied in any system. Here, we studied the in vivo interaction of U1-70K with an SR protein (SR45) and the mobility of the U1-70K/SR protein complex using bimolecular fluorescence complementation (BiFC) and fluorescence recovery after photobleaching (FRAP). Our results show that U1-70K exchanges between speckles and the nucleoplasmic pool very rapidly and that this exchange is sensitive to ongoing transcription and phosphorylation. BiFC analyses showed that U1-70K and SR45 interacted primarily in speckles and that this interaction is mediated by the RS1 or RS2 domain of SR45. FRAP analyses showed considerably slower recovery of the SR45/U1-70K complex than either protein alone indicating that SR45/U1-70K complexes remain in the speckles for a longer duration. Furthermore, FRAP analyses with SR45/U1-70K complex in the presence of inhibitors of phosphorylation did not reveal any significant change compared to control cells, suggesting that the mobility of the complex is not affected by the status of protein phosphorylation. These results indicate that U1-70K, like SR splicing factors, moves rapidly in the nucleus ensuring its availability at various sites of splicing. Furthermore, although it appears that U1-70K moves by diffusion its mobility is regulated by phosphorylation and transcription.
Conflict of interest statement
Figures



References
-
- Reddy AS. Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu Rev Plant Biol. 2007;58:267–294. - PubMed
-
- Reddy ASN. Nuclear Pre-mRNA Splicing in Plants. Critical Reviews in Plant Sciences. 2001;20:523–571.
-
- Graveley BR. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 2001;17:100–107. - PubMed
-
- Ast G. How did alternative splicing evolve? Nat Rev Genet. 2004;5:773–782. - PubMed
-
- Maniatis T, Tasic B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418:236–243. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

