A single Banana streak virus integration event in the banana genome as the origin of infectious endogenous pararetrovirus
- PMID: 18417582
- PMCID: PMC2447048
- DOI: 10.1128/JVI.00212-08
A single Banana streak virus integration event in the banana genome as the origin of infectious endogenous pararetrovirus
Abstract
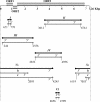
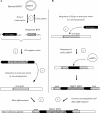
Sequencing of plant nuclear genomes reveals the widespread presence of integrated viral sequences known as endogenous pararetroviruses (EPRVs). Banana is one of the three plant species known to harbor infectious EPRVs. Musa balbisiana carries integrated copies of Banana streak virus (BSV), which are infectious by releasing virions in interspecific hybrids. Here, we analyze the organization of the EPRV of BSV Goldfinger (BSGfV) present in the wild diploid M. balbisiana cv. Pisang Klutuk Wulung (PKW) revealed by the study of Musa bacterial artificial chromosome resources and interspecific genetic cross. cv. PKW contains two similar EPRVs of BSGfV. Genotyping of these integrants and studies of their segregation pattern show an allelic insertion. Despite the fact that integrated BSGfV has undergone extensive rearrangement, both EPRVs contain the full-length viral genome. The high degree of sequence conservation between the integrated and episomal form of the virus indicates a recent integration event; however, only one allele is infectious. Analysis of BSGfV EPRV segregation among an F1 population from an interspecific genetic cross revealed that these EPRV sequences correspond to two alleles originating from a single integration event. We describe here for the first time the full genomic and genetic organization of the two EPRVs of BSGfV present in cv. PKW in response to the challenge facing both scientists and breeders to identify and generate genetic resources free from BSV. We discuss the consequences of this unique host-pathogen interaction in terms of genetic and genomic plant defenses versus strategies of infectious BSGfV EPRVs.
Figures




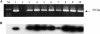


References
-
- Altschul, S., W. Gish, W. Miller, E. Myers, and D. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215403-410. - PubMed
-
- Amedeo, P., Y. Habu, K. Afsar, O. M. Scheid, and J. Paszkowski. 2000. Disruption of the plant gene MOM releases transcriptional silencing of methylated genes. Nature 405203-206. - PubMed
-
- Berriman, M., and K. Rutherford. 2003. Viewing and annotating sequence data with Artemis. Brief Bioinform. 4124-132. - PubMed
-
- Capy, P., G. Gasperi, C. Biemont, and C. Bazin. 2000. Stress and transposable elements: co-evolution or useful parasites? Heredity 85101-106. - PubMed
-
- Cocquet, J., A. Chong, G. L. Zhang, and R. A. Veitia. 2006. Reverse transcriptase template switching and false alternative transcripts. Genomics 88127-131. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

