Rapid sequence change and geographical spread of human parvovirus B19: comparison of B19 virus evolution in acute and persistent infections
- PMID: 18417586
- PMCID: PMC2447064
- DOI: 10.1128/JVI.00471-08
Rapid sequence change and geographical spread of human parvovirus B19: comparison of B19 virus evolution in acute and persistent infections
Abstract
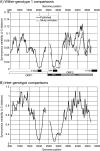
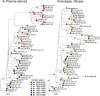
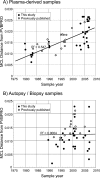
Parvovirus B19 is a common human pathogen maintained by horizontal transmission between acutely infected individuals. However, B19 virus can also be detected in tissues throughout the life of the host, although little is understood about the nature of such persistence. In the current study, we created large VP1/2 sequence data sets of plasma- and tissue (autopsy)-derived variants of B19 virus with known sample dates to compare the rates of sequence change in exogenous virus populations with those in persistently infected individuals. By using linear regression and likelihood-based methods (such as the BEAST program), we found that plasma-derived B19 virus showed a substitution rate of 4 x 10(-4) and an unconstrained (synonymous)-substitution rate of 18 x 10(-4) per site per year, several times higher than previously estimated and within the range of values for mammalian RNA viruses. The underlying high mutation frequency implied by these substitution rates may enable rapid adaptive changes that are more commonly ascribed to RNA virus populations. These revised estimates predict that the last common ancestor for currently circulating genotype 1 variants of B19 virus existed around 1956 to 1959, fitting well with previous analyses of the B19 virus "bioportfolio" that support a complete cessation of genotype 2 infections and their replacement by genotype 1 infections in the 1960s. In contrast, the evolution of B19 virus amplified from tissue samples was best modeled by using estimated dates of primary infection rather than sample dates, consistent with slow or absent sequence change during persistence. Determining what epidemiological or biological factors led to such a complete and geographically extensive population replacement over this short period is central to further understanding the nature of parvovirus evolution.
Figures
References
-
- Barros de Freitas, R., E. L. Durigon, D. S. Oliveira, C. M. Romano, M. R. Castro de Freitas, A. C. Linhares, F. L. Melo, L. Walshkeller, M. L. Barbosa, E. M. Huatuco, E. C. Holmes, and P. M. Zanotto. 2007. The “pressure pan” evolution of human erythrovirus B19 in the Amazon, Brazil. Virology 369281-287. - PubMed
-
- Brown, K. E., S. M. Anderson, and N. S. Young. 1993. Erythrocyte P antigen: cellular receptor for B19 parvovirus. Science 262114-117. - PubMed
-
- Brown, K. E., and P. Simmonds. 2007. Parvoviruses and blood transfusion. Transfusion 471745-1750. - PubMed
-
- Brown, T., A. Anand, L. D. Ritchie, J. P. Clewley, and T. M. Reid. 1984. Intrauterine parvovirus infection associated with hydrops fetalis. Lancet ii1033-1034. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

