Analysis of the yeast kinome reveals a network of regulated protein localization during filamentous growth
- PMID: 18417610
- PMCID: PMC2441683
- DOI: 10.1091/mbc.e07-11-1199
Analysis of the yeast kinome reveals a network of regulated protein localization during filamentous growth
Abstract
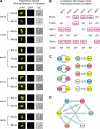
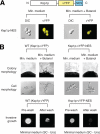
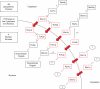
The subcellular distribution of kinases and other signaling proteins is regulated in response to cellular cues; however, the extent of this regulation has not been investigated for any gene set in any organism. Here, we present a systematic analysis of protein kinases in the budding yeast, screening for differential localization during filamentous growth. Filamentous growth is an important stress response involving mitogen-activated protein kinase and cAMP-dependent protein kinase signaling modules, wherein yeast cells form interconnected and elongated chains. Because standard strains of yeast are nonfilamentous, we constructed a unique set of 125 kinase-yellow fluorescent protein chimeras in the filamentous Sigma1278b strain for this study. In total, we identified six cytoplasmic kinases (Bcy1p, Fus3p, Ksp1p, Kss1p, Sks1p, and Tpk2p) that localize predominantly to the nucleus during filamentous growth. These kinases form part of an interdependent, localization-based regulatory network: deletion of each individual kinase, or loss of kinase activity, disrupts the nuclear translocation of at least two other kinases. In particular, this study highlights a previously unknown function for the kinase Ksp1p, indicating the essentiality of its nuclear translocation during yeast filamentous growth. Thus, the localization of Ksp1p and the other kinases identified here is tightly controlled during filamentous growth, representing an overlooked regulatory component of this stress response.
Figures




References
-
- Bao M. Z., Schwartz M. A., Cantin G. T., Yates J. R., Madhani H. Pheromone-dependent destruction of the Tec1 transcription factor is required for MAP kinase signaling specificity in yeast. Cell. 2004;119:991–1000. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

