Genetic-linkage mapping of complex hereditary disorders to a whole-genome molecular-interaction network
- PMID: 18417725
- PMCID: PMC2493404
- DOI: 10.1101/gr.075622.107
Genetic-linkage mapping of complex hereditary disorders to a whole-genome molecular-interaction network
Abstract
Common hereditary neurodevelopmental disorders such as autism, bipolar disorder, and schizophrenia are most likely both genetically multifactorial and heterogeneous. Because of these characteristics traditional methods for genetic analysis fail when applied to such diseases. To address the problem we propose a novel probabilistic framework that combines the standard genetic linkage formalism with whole-genome molecular-interaction data to predict pathways or networks of interacting genes that contribute to common heritable disorders. We apply the model to three large genotype-phenotype data sets, identify a small number of significant candidate genes for autism (24), bipolar disorder (21), and schizophrenia (25), and predict a number of gene targets likely to be shared among the disorders.
Figures


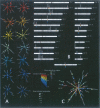
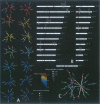

References
-
- Aerts S., Lambrechts D., Maity S., Van Loo P., Coessens B., De Smet F., Tranchevent L.C., De Moor B., Marynen P., Hassan B., et al. Gene prioritization through genomic data fusion. Nat. Biotechnol. 2006;24:537–544. - PubMed
-
- Akande E., Xenitidis K., Roberston M.D., Gorman J.M. Autism or schizophrenia: A diagnostic dilemma in adults with intellectual disabilities. J. Psychiatr. Pract. 2004;10:190–195. - PubMed
-
- Ars E., Serra E., Garcia J., Kruyer H., Gaona A., Lazaro C., Estivill X. Mutations affecting mRNA splicing are the most common molecular defects in patients with neurofibromatosis type 1. Hum. Mol. Genet. 2000;9:237–247. - PubMed
-
- Bacchelli E., Blasi F., Biondolillo M., Lamb J.A., Bonora E., Barnby G., Parr J., Beyer K.S., Klauck S.M., Poustka A., et al. Screening of nine candidate genes for autism on chromosome 2q reveals rare nonsynonymous variants in the cAMP-GEFII gene. Mol. Psychiatry. 2003;8:916–924. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
