Novel TCF-binding sites specify transcriptional repression by Wnt signalling
- PMID: 18418383
- PMCID: PMC2396397
- DOI: 10.1038/emboj.2008.80
Novel TCF-binding sites specify transcriptional repression by Wnt signalling
Abstract
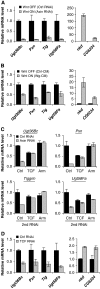
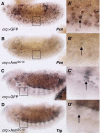
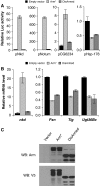
Both transcriptional activation and repression have essential functions in maintaining proper spatial and temporal control of gene expression. Although Wnt signalling is often associated with gene activation, we have identified several directly repressed targets of Wnt signalling in Drosophila. Here, we explore how individual Wnt target genes are specified for signal-induced activation or repression. Similar to activation, repression required binding of Armadillo (Arm) to the N terminus of TCF. However, TCF/Arm mediated repression by binding to DNA motifs that are markedly different from typical TCF-binding sites. Conversion of the novel motifs to standard TCF-binding sites reversed the mode of regulation, resulting in Wnt-mediated activation instead of repression. A mutant form of Arm defective in activation was still functional for repression, indicating that distinct domains of the protein are required for each activity. This study suggests that the sequence of TCF-binding sites allosterically regulates the TCF/Arm complex to effect either transcriptional activation or repression.
Figures




References
-
- Alfonso TB, Jones BW (2002) gcm2 promotes glial cell differentiation and is required with glial cells missing for macrophage development in Drosophila. Dev Biol 248: 369–383 - PubMed
-
- Barolo S (2006) Transgenic Wnt/TCF pathway reporters: all you need is Lef? Oncogene 25: 7505–7511 - PubMed
-
- Barolo S, Posakony JW (2002) Three habits of highly effective signaling pathways: principles of transcriptional control by developmental cell signaling. Genes Dev 16: 1167–1181 - PubMed
-
- Brand AH, Perrimon N (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

