Optimizing glycosyltransferase specificity via "hot spot" saturation mutagenesis presents a catalyst for novobiocin glycorandomization
- PMID: 18420146
- PMCID: PMC2813856
- DOI: 10.1016/j.chembiol.2008.02.017
Optimizing glycosyltransferase specificity via "hot spot" saturation mutagenesis presents a catalyst for novobiocin glycorandomization
Abstract
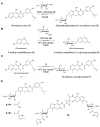
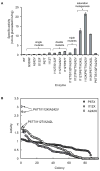
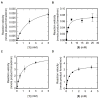
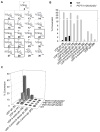
A comprehensive two-phase "hot spot" saturation mutagenesis strategy for the rapid evolution of glycosyltransferase (GT) specificity for nonnatural acceptors is described. Specifically, the application of a high-throughput screen (based on the fluorescent acceptor umbelliferone) was used to identify key amino acid hot spots that contribute to GT proficiency and/or promiscuity. Saturation mutagenesis of the corresponding hot spots facilitated the utilization of a lower-throughput screen to provide OleD prodigy capable of efficiently glycosylating the nonnatural acceptor novobiocic acid with an array of unique sugars. Incredibly, even in the absence of a high-throughput screen for novobiocic acid glycosylation, this approach rapidly led to improvements in the desired catalytic activity of several hundred-fold.
Figures

Comment in
-
Sweetly expanding enzymatic glycodiversification.Chem Biol. 2008 Apr;15(4):307-8. doi: 10.1016/j.chembiol.2008.03.009. Chem Biol. 2008. PMID: 18420135
References
-
- Thorson JS, Vogt T. Glycosylated natural products. In: Wong C-H, editor. Carbohydrate-Based Drug Discovery. Weinheim: Wiley-VCH; 2002. pp. 685–712.
-
- Weymouth-Wilson AC. The role of carbohydrates in biologically active natural products. Nat Prod Rep. 1997;14:99–110. - PubMed
-
- Ahmed A, Peters NR, Fitzgerald MK, Watson JA, Jr, Hoffmann FM, Thorson JS. Colchicine glycorandomization influences cytotoxicity and mechanism of action. J Am Chem Soc. 2006;128:14224–14225. - PubMed
-
- Langenhan JM, Griffith BR, Thorson JS. Neoglycorandomization and chemoenzymatic glycorandomization: two complementary tools for natural product diversification. J Nat Prod. 2005;68:1696–1711. - PubMed
-
- Griffith BR, Langenhan JM, Thorson JS. ‘Sweetening’ natural products via glycorandomization. Curr Opin Biotechnol. 2005;16:622–630. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

