The MUT9p kinase phosphorylates histone H3 threonine 3 and is necessary for heritable epigenetic silencing in Chlamydomonas
- PMID: 18420823
- PMCID: PMC2359792
- DOI: 10.1073/pnas.0711310105
The MUT9p kinase phosphorylates histone H3 threonine 3 and is necessary for heritable epigenetic silencing in Chlamydomonas
Abstract
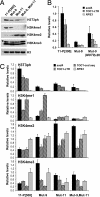
Changes in chromatin organization are emerging as key regulators in nearly every aspect of DNA-templated metabolism in eukaryotes. Histones undergo many, largely reversible, posttranslational modifications that affect chromatin structure. Some modifications, such as trimethylation of histone H3 on Lys 4 (H3K4me3), correlate with transcriptional activation, whereas others, such as methylation of histone H3 on Lys 27 (H3K27me), are associated with silent chromatin. Posttranslational histone modifications may also be involved in the inheritance of chromatin states. Histone phosphorylation has been implicated in a variety of cellular processes but, because of the dynamic nature of this modification, its potential role in long-term gene silencing has remained relatively unexplored. We report here that a Chlamydomonas reinhardtii mutant defective in a Ser/Thr protein kinase (MUT9p), which phosphorylates histones H3 and H2A, shows deficiencies in the heritable repression of transgenes and transposons. Moreover, based on chromatin immunoprecipitation analyses, phosphorylated H3T3 (H3T3ph) and monomethylated H3K4 (H3K4me1) are inversely correlated with di/trimethylated H3K4 and associate preferentially with silenced transcription units. Conversely, the loss of those marks in mutant strains correlates with the transcriptional reactivation of transgenes and transposons. Our results suggest that H3T3ph and H3K4me1 function as reinforcing epigenetic marks for the silencing of euchromatic loci in Chlamydomonas.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. - PubMed
-
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. - PubMed
-
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. - PubMed
-
- Latham JA, Dent SY. Cross-regulation of histone modifications. Nat Struct Mol Biol. 2007;14:1017–1024. - PubMed
-
- Henderson IR, Jacobsen SE. Epigenetic inheritance in plants. Nature. 2007;447:418–424. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

