The chemical genomic portrait of yeast: uncovering a phenotype for all genes
- PMID: 18420932
- PMCID: PMC2794835
- DOI: 10.1126/science.1150021
The chemical genomic portrait of yeast: uncovering a phenotype for all genes
Abstract
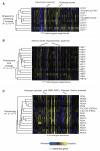
Genetics aims to understand the relation between genotype and phenotype. However, because complete deletion of most yeast genes ( approximately 80%) has no obvious phenotypic consequence in rich medium, it is difficult to study their functions. To uncover phenotypes for this nonessential fraction of the genome, we performed 1144 chemical genomic assays on the yeast whole-genome heterozygous and homozygous deletion collections and quantified the growth fitness of each deletion strain in the presence of chemical or environmental stress conditions. We found that 97% of gene deletions exhibited a measurable growth phenotype, suggesting that nearly all genes are essential for optimal growth in at least one condition.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

