Analysis of chimpanzee history based on genome sequence alignments
- PMID: 18421364
- PMCID: PMC2278377
- DOI: 10.1371/journal.pgen.1000057
Analysis of chimpanzee history based on genome sequence alignments
Abstract
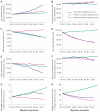
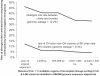
Population geneticists often study small numbers of carefully chosen loci, but it has become possible to obtain orders of magnitude for more data from overlaps of genome sequences. Here, we generate tens of millions of base pairs of multiple sequence alignments from combinations of three western chimpanzees, three central chimpanzees, an eastern chimpanzee, a bonobo, a human, an orangutan, and a macaque. Analysis provides a more precise understanding of demographic history than was previously available. We show that bonobos and common chimpanzees were separated approximately 1,290,000 years ago, western and other common chimpanzees approximately 510,000 years ago, and eastern and central chimpanzees at least 50,000 years ago. We infer that the central chimpanzee population size increased by at least a factor of 4 since its separation from western chimpanzees, while the western chimpanzee effective population size decreased. Surprisingly, in about one percent of the genome, the genetic relationships between humans, chimpanzees, and bonobos appear to be different from the species relationships. We used PCR-based resequencing to confirm 11 regions where chimpanzees and bonobos are not most closely related. Study of such loci should provide information about the period of time 5-7 million years ago when the ancestors of humans separated from those of the chimpanzees.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Groves C. Washington D.C.: Smithsonian Institution Press; 2001. Primate taxonomy. p. 350.
-
- Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

