An unbiased systems genetics approach to mapping genetic loci modulating susceptibility to severe streptococcal sepsis
- PMID: 18421376
- PMCID: PMC2277464
- DOI: 10.1371/journal.ppat.1000042
An unbiased systems genetics approach to mapping genetic loci modulating susceptibility to severe streptococcal sepsis
Abstract
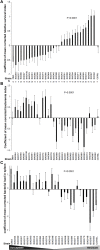
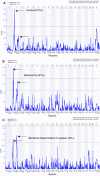
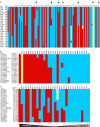
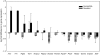
Striking individual differences in severity of group A streptococcal (GAS) sepsis have been noted, even among patients infected with the same bacterial strain. We had provided evidence that HLA class II allelic variation contributes significantly to differences in systemic disease severity by modulating host responses to streptococcal superantigens. Inasmuch as the bacteria produce additional virulence factors that participate in the pathogenesis of this complex disease, we sought to identify additional gene networks modulating GAS sepsis. Accordingly, we applied a systems genetics approach using a panel of advanced recombinant inbred mice. By analyzing disease phenotypes in the context of mice genotypes we identified a highly significant quantitative trait locus (QTL) on Chromosome 2 between 22 and 34 Mb that strongly predicts disease severity, accounting for 25%-30% of variance. This QTL harbors several polymorphic genes known to regulate immune responses to bacterial infections. We evaluated candidate genes within this QTL using multiple parameters that included linkage, gene ontology, variation in gene expression, cocitation networks, and biological relevance, and identified interleukin1 alpha and prostaglandin E synthases pathways as key networks involved in modulating GAS sepsis severity. The association of GAS sepsis with multiple pathways underscores the complexity of traits modulating GAS sepsis and provides a powerful approach for analyzing interactive traits affecting outcomes of other infectious diseases.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Sumby P, Porcella SF, Madrigal AG, Barbian KD, Virtaneva K, et al. Evolutionary origin and emergence of a highly successful clone of serotype M1 group a Streptococcus involved multiple horizontal gene transfer events. J Infect Dis. 2005;192:771–782. - PubMed
-
- Walker MJ, Hollands A, Sanderson-Smith ML, Cole JN, Kirk JK, et al. DNase Sda1 provides selection pressure for a switch to invasive group A streptococcal infection. Nat Med. 2007;13:981–985. - PubMed
-
- Low DE, Schwartz B, McGeer A. Washington, D.C: American Society for Microbiology Press; 1997. The reemergence of severe group A streptococcal disease: an evolutionary perspetive. pp. 93–123.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

