Diversification and adaptive sequence evolution of Caenorhabditis lysozymes (Nematoda: Rhabditidae)
- PMID: 18423043
- PMCID: PMC2383907
- DOI: 10.1186/1471-2148-8-114
Diversification and adaptive sequence evolution of Caenorhabditis lysozymes (Nematoda: Rhabditidae)
Abstract
Background: Lysozymes are important model enzymes in biomedical research with a ubiquitous taxonomic distribution ranging from phages up to plants and animals. Their main function appears to be defence against pathogens, although some of them have also been implicated in digestion. Whereas most organisms have only few lysozyme genes, nematodes of the genus Caenorhabditis possess a surprisingly large repertoire of up to 15 genes.
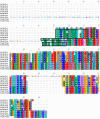
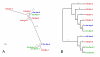
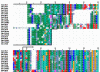
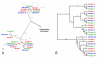
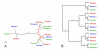
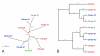
Results: We used phylogenetic inference and sequence analysis tools to assess the evolution of lysozymes from three congeneric nematode species, Caenorhabditis elegans, C. briggsae, and C. remanei. Their lysozymes fall into three distinct clades, one belonging to the invertebrate-type and the other two to the protist-type lysozymes. Their diversification is characterised by (i) ancestral gene duplications preceding species separation followed by maintenance of genes, (ii) ancestral duplications followed by gene loss in some of the species, and (iii) recent duplications after divergence of species. Both ancestral and recent gene duplications are associated in several cases with signatures of adaptive sequence evolution, indicating that diversifying selection contributed to lysozyme differentiation. Current data strongly suggests that genetic diversity translates into functional diversity.
Conclusion: Gene duplications are a major source of evolutionary innovation. Our analysis provides an evolutionary framework for understanding the diversification of lysozymes through gene duplication and subsequent differentiation. This information is expected to be of major value in future analysis of lysozyme function and in studies of the dynamics of evolution by gene duplication.
Figures


References
-
- Jollès P. In: Lysozymes: Model enzymes in biochemistry and biology. Jollès P, editor. Basel, Switzerland , Birkhäuser Verlag; 1996. From the discovery of lysozyme to the characterization of several lysozyme families; pp. 3–5. - PubMed
-
- Prager EM, Jollès P. In: Lysozymes: Model enzymes in biochemistry and biology. Jollès P, editor. Basel, Switzerland , Birkhäuser Verlag; 1996. Animal lysozymes c and g: An overview; pp. 9–31. - PubMed
-
- Fastrez J. In: Lysozymes: Model enzymes in biochemistry and biology. Jollès P, editor. Basel, Switzerland , Birkhäuser Verlag; 1996. Phage lysozymes; pp. 35–64.
-
- Höltje JV. In: Lysozymes: Model enzymes in biochemistry and biology. Jollès P, editor. Basel, Switzerland , Birkhäuser Verlag; 1996. Bacterial lysozymes; pp. 65–74.
-
- Hultmark D. In: Lysozymes: Model enzymes in biochemistry and biology. Jollès P, editor. Basel, Switzerland , Birkhäuser Verlag; 1996. Insect lysozymes; pp. 87–101.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

