Genome-wide subcellular localization of putative outer membrane and extracellular proteins in Leptospira interrogans serovar Lai genome using bioinformatics approaches
- PMID: 18423054
- PMCID: PMC2387172
- DOI: 10.1186/1471-2164-9-181
Genome-wide subcellular localization of putative outer membrane and extracellular proteins in Leptospira interrogans serovar Lai genome using bioinformatics approaches
Erratum in
- BMC Genomics. 2008;9:602
Abstract
Background: In bacterial pathogens, both cell surface-exposed outer membrane proteins and proteins secreted into the extracellular environment play crucial roles in host-pathogen interaction and pathogenesis. Considerable efforts have been made to identify outer membrane (OM) and extracellular (EX) proteins produced by Leptospira interrogans, which may be used as novel targets for the development of infection markers and leptospirosis vaccines.
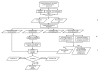
Result: In this study we used a novel computational framework based on combined prediction methods with deduction concept to identify putative OM and EX proteins encoded by the Leptospira interrogans genome. The framework consists of the following steps: (1) identifying proteins homologous to known proteins in subcellular localization databases derived from the "consensus vote" of computational predictions, (2) incorporating homology based search and structural information to enhance gene annotation and functional identification to infer the specific structural characters and localizations, and (3) developing a specific classifier for cytoplasmic proteins (CP) and cytoplasmic membrane proteins (CM) using Linear discriminant analysis (LDA). We have identified 114 putative EX and 63 putative OM proteins, of which 41% are conserved or hypothetical proteins containing sequence and/or protein folding structures similar to those of known EX and OM proteins.
Conclusion: Overall results derived from the combined computational analysis correlate with the available experimental evidence. This is the most extensive in silico protein subcellular localization identification to date for Leptospira interrogans serovar Lai genome that may be useful in protein annotation, discovery of novel genes and understanding the biology of Leptospira.
Figures
Similar articles
-
In silico and microarray-based genomic approaches to identifying potential vaccine candidates against Leptospira interrogans.BMC Genomics. 2006 Nov 16;7:293. doi: 10.1186/1471-2164-7-293. BMC Genomics. 2006. PMID: 17109759 Free PMC article.
-
Genetic diversity among major endemic strains of Leptospira interrogans in China.BMC Genomics. 2007 Jul 1;8:204. doi: 10.1186/1471-2164-8-204. BMC Genomics. 2007. PMID: 17603913 Free PMC article.
-
Whole-genome analysis of Leptospira interrogans to identify potential vaccine candidates against leptospirosis.FEMS Microbiol Lett. 2005 Mar 15;244(2):305-13. doi: 10.1016/j.femsle.2005.02.004. FEMS Microbiol Lett. 2005. PMID: 15766783
-
New insights into the putative role of leucine-rich repeat proteins of Leptospira interrogans and their participation in host cell invasion: an in silico analysis.Front Cell Infect Microbiol. 2024 Dec 13;14:1492352. doi: 10.3389/fcimb.2024.1492352. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39735260 Free PMC article. Review.
-
Technological advances in the molecular biology of Leptospira.J Mol Microbiol Biotechnol. 2000 Oct;2(4):455-62. J Mol Microbiol Biotechnol. 2000. PMID: 11075918 Review.
Cited by
-
Bioinformatic Exploration of Metal-Binding Proteome of Zoonotic Pathogen Orientia tsutsugamushi.Front Genet. 2019 Sep 24;10:797. doi: 10.3389/fgene.2019.00797. eCollection 2019. Front Genet. 2019. PMID: 31608099 Free PMC article.
-
Leptospira: the dawn of the molecular genetics era for an emerging zoonotic pathogen.Nat Rev Microbiol. 2009 Oct;7(10):736-47. doi: 10.1038/nrmicro2208. Nat Rev Microbiol. 2009. PMID: 19756012 Free PMC article. Review.
-
Challenges for the development of a universal vaccine against leptospirosis revealed by the evaluation of 22 vaccine candidates.Front Cell Infect Microbiol. 2022 Oct 7;12:940966. doi: 10.3389/fcimb.2022.940966. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36275031 Free PMC article.
-
Insight into the Structure, Functions, and Dynamics of the Leptospira Outer Membrane Proteins with the Pathogenicity.Membranes (Basel). 2022 Mar 7;12(3):300. doi: 10.3390/membranes12030300. Membranes (Basel). 2022. PMID: 35323775 Free PMC article. Review.
-
A putative regulatory genetic locus modulates virulence in the pathogen Leptospira interrogans.Infect Immun. 2014 Jun;82(6):2542-52. doi: 10.1128/IAI.01803-14. Epub 2014 Mar 31. Infect Immun. 2014. PMID: 24686063 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

