Porphyrin binding and distortion and substrate specificity in the ferrochelatase reaction: the role of active site residues
- PMID: 18423489
- PMCID: PMC2852141
- DOI: 10.1016/j.jmb.2008.03.040
Porphyrin binding and distortion and substrate specificity in the ferrochelatase reaction: the role of active site residues
Abstract
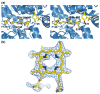
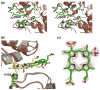
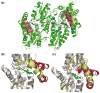
The specific insertion of a divalent metal ion into tetrapyrrole macrocycles is catalyzed by a group of enzymes called chelatases. Distortion of the tetrapyrrole has been proposed to be an important component of the mechanism of metallation. We present the structures of two different inhibitor complexes: (1) N-methylmesoporphyrin (N-MeMP) with the His183Ala variant of Bacillus subtilis ferrochelatase; (2) the wild-type form of the same enzyme with deuteroporphyrin IX 2,4-disulfonic acid dihydrochloride (dSDP). Analysis of the structures showed that only one N-MeMP isomer out of the eight possible was bound to the protein and it was different from the isomer that was earlier found to bind to the wild-type enzyme. A comparison of the distortion of this porphyrin with other porphyrin complexes of ferrochelatase and a catalytic antibody with ferrochelatase activity using normal-coordinate structural decomposition reveals that certain types of distortion are predominant in all these complexes. On the other hand, dSDP, which binds closer to the protein surface compared to N-MeMP, does not undergo any distortion upon binding to the protein, underscoring that the position of the porphyrin within the active site pocket is crucial for generating the distortion required for metal insertion. In addition, in contrast to the wild-type enzyme, Cu(2+)-soaking of the His183Ala variant complex did not show any traces of porphyrin metallation. Collectively, these results provide new insights into the role of the active site residues of ferrochelatase in controlling stereospecificity, distortion and metallation.
Figures
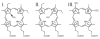

References
-
- Fraústo da Silva RJJ, Williams RJP. The Biological Chemistry of the Elements. Vol. 2. Oxford University Press; Oxford, UK: 2001. Haem iron: coupled redox reactions; pp. 370–399.
-
- Dailey HA, Dailey TA. Ferrochelatase. In: Kadish KM, Smith KM, Guilard R, editors. Porphyrin Handbook: The Iron and Cobalt Pigments: Biosynthesis, Structure and Degradation. Vol. 12. Academic Press; San Diego, CA: 2003. pp. 93–122.
-
- Willows RD, Hansson M. Mechanism, structure, and regulation of magnesium chelatase. In: Kadish KM, Smith KM, Guilard R, editors. Porphyrin Handbook: Chlorophylls and Bilins: Biosynthesis, Synthesis and Degradation. Vol. 13. Academic Press; San Diego, CA: 2003. pp. 1–47.
-
- Schubert HL, Raux E, Wilson KS, Warren MJ. Common chelatase design in the branched tetrapyrrole pathways of heme and anaerobic cobalamin synthesis. Biochemistry. 1999;38:10660–10669. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

