UpaG, a new member of the trimeric autotransporter family of adhesins in uropathogenic Escherichia coli
- PMID: 18424525
- PMCID: PMC2446758
- DOI: 10.1128/JB.00122-08
UpaG, a new member of the trimeric autotransporter family of adhesins in uropathogenic Escherichia coli
Abstract
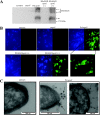
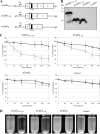
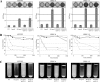
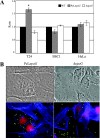
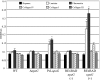
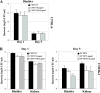
The ability of Escherichia coli to colonize both intestinal and extraintestinal sites is driven by the presence of specific virulence factors, among which are the autotransporter (AT) proteins. Members of the trimeric AT adhesin family are important virulence factors for several gram-negative pathogens and mediate adherence to eukaryotic cells and extracellular matrix (ECM) proteins. In this study, we characterized a new trimeric AT adhesin (UpaG) from uropathogenic E. coli (UPEC). Molecular analysis of UpaG revealed that it is translocated to the cell surface and adopts a multimeric conformation. We demonstrated that UpaG is able to promote cell aggregation and biofilm formation on abiotic surfaces in CFT073 and various UPEC strains. In addition, UpaG expression resulted in the adhesion of CFT073 to human bladder epithelial cells, with specific affinity to fibronectin and laminin. Prevalence analysis revealed that upaG is strongly associated with E. coli strains from the B2 and D phylogenetic groups, while deletion of upaG had no significant effect on the ability of CFT073 to colonize the mouse urinary tract. Thus, UpaG is a novel trimeric AT adhesin from E. coli that mediates aggregation, biofilm formation, and adhesion to various ECM proteins.
Figures

Similar articles
-
Molecular characterization of the EhaG and UpaG trimeric autotransporter proteins from pathogenic Escherichia coli.Appl Environ Microbiol. 2012 Apr;78(7):2179-89. doi: 10.1128/AEM.06680-11. Epub 2012 Jan 27. Appl Environ Microbiol. 2012. PMID: 22286983 Free PMC article.
-
Functional analysis of antigen 43 in uropathogenic Escherichia coli reveals a role in long-term persistence in the urinary tract.Infect Immun. 2007 Jul;75(7):3233-44. doi: 10.1128/IAI.01952-06. Epub 2007 Apr 9. Infect Immun. 2007. PMID: 17420234 Free PMC article.
-
Molecular characterization of UpaB and UpaC, two new autotransporter proteins of uropathogenic Escherichia coli CFT073.Infect Immun. 2012 Jan;80(1):321-32. doi: 10.1128/IAI.05322-11. Epub 2011 Sep 19. Infect Immun. 2012. PMID: 21930758 Free PMC article.
-
Covert operations of uropathogenic Escherichia coli within the urinary tract.Traffic. 2005 Jan;6(1):18-31. doi: 10.1111/j.1600-0854.2004.00251.x. Traffic. 2005. PMID: 15569242 Free PMC article. Review.
-
Adhesins and invasins of pathogenic Escherichia coli.Int J Med Microbiol. 2005 Oct;295(6-7):471-8. doi: 10.1016/j.ijmm.2005.07.001. Int J Med Microbiol. 2005. PMID: 16238021 Review.
Cited by
-
Adh enhances Actinobacillus pleuropneumoniae pathogenicity by binding to OR5M11 and activating p38 which induces apoptosis of PAMs and IL-8 release.Sci Rep. 2016 Apr 5;6:24058. doi: 10.1038/srep24058. Sci Rep. 2016. PMID: 27046446 Free PMC article.
-
Genome-wide mapping of cystitis due to Streptococcus agalactiae and Escherichia coli in mice identifies a unique bladder transcriptome that signifies pathogen-specific antimicrobial defense against urinary tract infection.Infect Immun. 2012 Sep;80(9):3145-60. doi: 10.1128/IAI.00023-12. Epub 2012 Jun 25. Infect Immun. 2012. PMID: 22733575 Free PMC article.
-
Adhesive threads of extraintestinal pathogenic Escherichia coli.Gut Pathog. 2009 Dec 10;1(1):22. doi: 10.1186/1757-4749-1-22. Gut Pathog. 2009. PMID: 20003270 Free PMC article.
-
In Vivo Role of Two-Component Regulatory Systems in Models of Urinary Tract Infections.Pathogens. 2023 Jan 10;12(1):119. doi: 10.3390/pathogens12010119. Pathogens. 2023. PMID: 36678467 Free PMC article. Review.
-
Muramidases found in the foregut microbiome of the Tammar wallaby can direct cell aggregation and biofilm formation.ISME J. 2011 Feb;5(2):341-50. doi: 10.1038/ismej.2010.116. Epub 2010 Jul 29. ISME J. 2011. PMID: 20668486 Free PMC article.
References
-
- Belda, E., A. Moya, and F. J. Silva. 2005. Genome rearrangement distances and gene order phylogeny in gamma-Proteobacteria. Mol. Biol. Evol. 221456-1467. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

