Characterization of the pattern of alphas1- and beta-casein breakdown and release of a bioactive peptide by a cell envelope proteinase from Lactobacillus delbrueckii subsp. lactis CRL 581
- PMID: 18424544
- PMCID: PMC2446539
- DOI: 10.1128/AEM.00247-08
Characterization of the pattern of alphas1- and beta-casein breakdown and release of a bioactive peptide by a cell envelope proteinase from Lactobacillus delbrueckii subsp. lactis CRL 581
Abstract
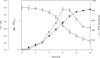
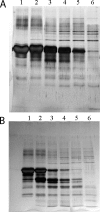
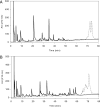
The cell envelope-associated proteinases (CEPs) of the lactobacilli have key roles in bacterial nutrition and contribute to the development of the organoleptic properties of fermented milk products as well, as they can release bioactive health-beneficial peptides from milk proteins. The influence of the peptide supply, carbohydrate source, and osmolites on the CEP activity of the cheese starter Lactobacillus delbrueckii subsp. lactis CRL 581 was investigated. The CEP activity levels were controlled by the peptide content of the growth medium. The maximum activity was observed in a basal minimal defined medium, whereas in the presence of Casitone, Casamino Acids, or yeast extract, the synthesis of CEP was inhibited 99-, 70-, and 68-fold, respectively. The addition of specific di- or tripeptides containing branched-chain amino acids, such as leucylleucine, prolylleucine, leucylglycylglycine, or leucylproline, to the growth medium negatively affected CEP activity, whereas dipeptides without branched-chain amino acids had no effect on the enzyme's production. The carbon source and osmolites did not affect CEP activity. The CEP of L. delbrueckii subsp. lactis CRL 581 exhibited a mixed-type CEP(I/III) variant caseinolytic specificity. Mass-spectrometric screening of the main peptide peaks isolated by reverse-phase high-pressure liquid chromatography allowed the identification of 33 and 32 peptides in the alpha(s1)- and beta-casein hydrolysates, respectively. By characterizing the peptide sequence in these hydrolysates, a pattern of alpha(s1)- and beta-casein breakdown was defined and is reported herein, this being the first report for a CEP of L. delbrueckii subsp. lactis. In this pattern, a series of potentially bioactive peptides (antihypertensive and phosphopeptides) which are encrypted within the precursor protein could be visualized.
Figures


Similar articles
-
Characterization of the mature cell surface proteinase of Lactobacillus delbrueckii subsp. lactis CRL 581.Appl Microbiol Biotechnol. 2015 May;99(10):4277-86. doi: 10.1007/s00253-014-6258-6. Epub 2014 Dec 9. Appl Microbiol Biotechnol. 2015. PMID: 25487890
-
Diversity in proteinase specificity of thermophilic lactobacilli as revealed by hydrolysis of dairy and vegetable proteins.Appl Microbiol Biotechnol. 2013 Sep;97(17):7831-44. doi: 10.1007/s00253-013-5037-0. Epub 2013 Jul 9. Appl Microbiol Biotechnol. 2013. PMID: 23832109
-
Release of the cell-envelope-associated proteinase of Lactobacillus delbrueckii subspecies lactis CRL 581 is dependent upon pH and temperature.J Agric Food Chem. 2009 Sep 23;57(18):8607-11. doi: 10.1021/jf901531q. J Agric Food Chem. 2009. PMID: 19754175
-
Original features of cell-envelope proteinases of Lactobacillus helveticus. A review.Int J Food Microbiol. 2011 Mar 15;146(1):1-13. doi: 10.1016/j.ijfoodmicro.2011.01.039. Epub 2011 Feb 2. Int J Food Microbiol. 2011. PMID: 21354644 Review.
-
Peptidomics methods for the identification of peptidase-substrate interactions.Curr Opin Chem Biol. 2013 Feb;17(1):83-9. doi: 10.1016/j.cbpa.2012.10.038. Epub 2013 Jan 15. Curr Opin Chem Biol. 2013. PMID: 23332665 Free PMC article. Review.
Cited by
-
Valorization of leftover green tea residues through conversion to bioactive peptides using probiotics-aided anaerobic digestion.Microb Biotechnol. 2023 Feb;16(2):418-431. doi: 10.1111/1751-7915.14155. Epub 2022 Oct 26. Microb Biotechnol. 2023. PMID: 36285915 Free PMC article.
-
Characterization of Cell-Envelope Proteinases from Two Lacticaseibacillus casei Strains Isolated from Parmigiano Reggiano Cheese.Biology (Basel). 2022 Jan 14;11(1):139. doi: 10.3390/biology11010139. Biology (Basel). 2022. PMID: 35053137 Free PMC article.
-
YebC, a putative transcriptional factor involved in the regulation of the proteolytic system of Lactobacillus.Sci Rep. 2017 Aug 17;7(1):8579. doi: 10.1038/s41598-017-09124-1. Sci Rep. 2017. PMID: 28819300 Free PMC article.
-
Identification of Antihypertensive Peptides Derived from Low Molecular Weight Casein Hydrolysates Generated during Fermentation by Bifidobacterium longum KACC 91563.Korean J Food Sci Anim Resour. 2015;35(6):738-47. doi: 10.5851/kosfa.2015.35.6.738. Epub 2015 Dec 31. Korean J Food Sci Anim Resour. 2015. PMID: 26877633 Free PMC article.
-
Diversity and Functional Properties of Lactic Acid Bacteria Isolated From Wild Fruits and Flowers Present in Northern Argentina.Front Microbiol. 2019 May 21;10:1091. doi: 10.3389/fmicb.2019.01091. eCollection 2019. Front Microbiol. 2019. PMID: 31164879 Free PMC article.
References
-
- Altermann, E., W. M. Russell, M. A. Azcarate-Peril, R. Barrangou, B. L. Buck, O. McAuliffe, N. Souther, A. Dobson, T. Duong, M. Callanan, S. Lick, A. Hamrick, R. Cano, and T. R. Klaenhammer. 2005. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc. Natl. Acad. Sci. USA 102:3906-3912. - PMC - PubMed
-
- Caira, S., P. Ferranti, M. Gatti, M. E. Fornasari, F. Barone, S. Lilla, G. Mucchetti, G. Picariello, L. Chianese, E. Neviani, and F. Addeo. 2003. Synthetic peptides as substrate for assaying the proteolytic activity of Lactobacillus helveticus. J. Dairy Res. 70:315-325. - PubMed
-
- den Hengst, C. D., S. A. van Hijum, J. M. Geurts, A. Nauta, J. Kok, and O. P. Kuipers. 2005. The Lactococcus lactis CodY regulon: identification of a conserved cis-regulatory element. J. Biol. Chem. 280:34332-34342. - PubMed
-
- Exterkate, F. A. 1990. Differences in short peptide-substrate cleavage by two cell-envelope-located serine proteinases of Lactococcus lactis subsp. cremoris are related to secondary binding specificity. Appl. Microbiol. Biotechnol. 33:401-406. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

