A role for ubiquitin in the spliceosome assembly pathway
- PMID: 18425143
- PMCID: PMC2737727
- DOI: 10.1038/nsmb.1401
A role for ubiquitin in the spliceosome assembly pathway
Abstract
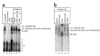
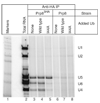
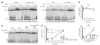
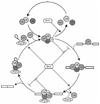
The spliceosome uses numerous strategies to regulate its function in mRNA maturation. Ubiquitin regulates many cellular processes, but its potential roles during splicing are unknown. We have developed a new strategy that reveals a direct role for ubiquitin in the dynamics of splicing complexes. A ubiquitin mutant (I44A) that can enter the conjugation pathway but is compromised in downstream functions diminishes splicing activity by reducing the levels of the U4/U6-U5 small nuclear ribonucleoprotein (snRNP). Similarly, an inhibitor of ubiquitin's protein-protein interactions, ubistatin A, reduces U4/U6-U5 triple snRNP levels in vitro. When ubiquitin interactions are blocked, ATP-dependent disassembly of purified U4/U6-U5 particles is accelerated, indicating a direct role for ubiquitin in repressing U4/U6 unwinding. Finally, we show that the conserved splicing factor Prp8 is ubiquitinated within purified triple snRNPs. These results reveal a previously unknown ubiquitin-dependent mechanism for controlling the pre-mRNA splicing pathway.
Figures



Comment in
-
Modifications target spliceosome dynamics.Nat Struct Mol Biol. 2008 May;15(5):426-8. doi: 10.1038/nsmb0508-426. Nat Struct Mol Biol. 2008. PMID: 18461042 No abstract available.
References
-
- Will CL, Lührmann R. Spliceosome structure and function. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2006. pp. 369–400.
-
- Staley JP, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. - PubMed
-
- Kuhn AN, Li Z, Brow DA. Splicing factor Prp8 governs U4/U6 RNA unwinding during activation of the spliceosome. Mol. Cell. 1999;3:65–75. - PubMed
-
- Misteli T. RNA splicing: what has phosphorylation got to do with it? Curr. Biol. 1999;9:R198–R200. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

