DNA watermarks: a proof of concept
- PMID: 18426578
- PMCID: PMC2375902
- DOI: 10.1186/1471-2199-9-40
DNA watermarks: a proof of concept
Abstract
Background: DNA-based watermarks are helpful tools to identify the unauthorized use of genetically modified organisms (GMOs) protected by patents. In silico analyses showed that in coding regions synonymous codons can be used to insert encrypted information into the genome of living organisms by using the DNA-Crypt algorithm.
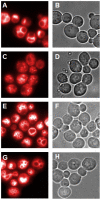
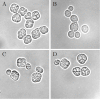
Results: We integrated an authenticating watermark in the Vam7 sequence. For our investigations we used a mutant Saccharomyces cerevisiae strain, called CG783, which has an amber mutation within the Vam7 sequence. The CG783 cells are unable to sporulate and in addition display an abnormal vacuolar morphology. Transformation of CG783 with pRS314 Vam7 leads to a phenotype very similar to the wildtype yeast strain CG781. The integrated watermark did not influence the function of Vam7 and the resulting phenotype of the CG783 cells transformed with pRS314 Vam7-TB shows no significant differences compared to the CG783 cells transformed with pRS314 Vam7.
Conclusion: From our experiments we conclude that the DNA watermarks produced by DNA-Crypt do not influence the translation from mRNA into protein. By analyzing the vacuolar morphology, growth rate and ability to sporulate we confirmed that the resulting Vam7 protein was functionally active.
Figures





References
-
- Gehani A, LaBean TH, Reif JH. DNA-based cryptography. Dimacs Series In Discrete Mathematics and Theoretical Computer Science. 2000;54:233–249.
-
- Wong PC, Wong KK, Foote H. Organic Data Memory Using the DNA Approach. Communications of the ACM. 2003;46
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

